Abstract
Syntaxin 6 (STX6), a soluble N-ethylmaleimide-sensitive factor-activating receptor protein, has formed an increasing part of cancer research. However, to the best of our knowledge, the role of STX6 in hepatocellular carcinoma (HCC) is still unclear. In the present study, data from multiple bioinformatics databases, including The Cancer Genome Atlas, Gene Expression Omnibus, Kaplan-Meier plotter, Tumor Immune Estimation Resource (TIMER) and Gene Expression Profiling Integrative Analysis (GEPIA2), and immunohistochemistry (IHC) were utilized to assess the role of STX6 in HCC. The results demonstrated that STX6 expression was upregulated in HCC tissues compared with normal tissues. STX6 expression was significantly associated with tumor size, Edmondson grade and α-fetoprotein (AFP) level. Furthermore, survival analysis demonstrated that high STX6 expression was significantly associated with poor prognosis in patients with HCC. Furthermore, assessment of the immune infiltrates demonstrated that CD163 expression was positively correlated with the STX6 level when analyzed using the TIMER and GEPIA2 databases. IHC results further demonstrated this association. Furthermore, compared with the typically used AFP, STX6 could have an improved diagnostic value in the diagnosis of HCC. In conclusion, STX6 expression was not only positively associated with poor prognosis but may also be involved in the immune inflammatory reaction in HCC. STX6 may become a potential therapeutic and diagnosis maker for patients with HCC.
Keywords: syntaxin 6, prognosis, immune infiltrates, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancy with the highest morbidity and mortality in numerous countries across the world (1). A number of treatments are used to treat HCC; however, the treatment effects are often limited, especially for advanced stage carcinomas (1,2). Therefore, it is urgent to further explore the carcinogenic targeting molecules of HCC to enhance prognosis and individualized treatments.
Syntaxin 6 (STX6) is a sensitive factor in the soluble n-ethylmaleimide receptor protein and has been reported to serve a role in Parkinson's disease (3,4). It has been reported to be expressed in brain, lung and kidney (5). Furthermore, STX6 is a vesicle transporter, which serves a vital role in intracellular protein transport and membrane structure changes (6). STX6 has been reported to be involved in tumorigenesis in multiple malignant tumors, including esophageal cancer (7), osteosarcoma (8) and renal cell carcinoma (9). Notably, a recent study reported that STX6 is a key factor in macrophages during the immune response in lipopolysaccharide-activated cells (10).
Macrophages are a major component of the inflammatory infiltrate in tumor (11,12). The high levels of tumor-associated macrophages (TAMs) are associated with poor prognosis in a range of tumors, including breast, gastric and colorectal carcinoma, and HCC (13–17). The macrophages are classified as M1 phenotype or M2 phenotype (18). It is now widely accepted that the M2 phenotype supports tumor growth (13–15,17,19). CD163 is widely reported as a scavenger receptor and is a highly specific marker of M2 macrophages (20,21). CD163 has been reported to be an anti-inflammatory molecule as it is mainly expressed by M2 macrophages at sites of inflammation (22). Interestingly, one study reported that STX6 is associated with increased cytokine secretion in activated macrophages (23). However, the mechanisms of STX6 immune infiltration in HCC remain unclear.
The present study analyzed the association between STX6 expression and clinical characteristics and prognosis of patients with HCC. Furthermore, the functions of STX6 in HCC and potential immune infiltration-related molecular mechanisms were assessed.
Materials and methods
Tissue sections
The tumor tissues and para-carcinoma tissue sections were collected between January 2014 and December 2016 and stored in the Human Resources Specimen Bank of the First Affiliated Hospital of Nanchang University (Nanchang, China). Samples were obtained from the Human Genetic Resources Center and Department of Pathology of The First Affiliated Hospital of Nanchang University (Nanchang, China) and examined independently by two pathologists. The patients included 76 males and 14 females between 27 and 81 years old (median 53). None of the patients had previously received other tumor surgery, chemotherapy, radiation therapy, or any other anticancer therapy. As this was a retrospective study, the requirement for informed consent was waived by the ethics committee. The present study was approved by the Clinical Medical Research Ethics Committee of the First Affiliated Hospital of Nanchang University (approval no. 202112020; Nanchang, China).
Immunohistochemistry (IHC)
To examine STX6 and CD163 expression in tumor tissues, paired paraffin-embedded tumor slices were obtained from the specimen bank. The sections were then incubated with anti-STX6 (1:100; cat. no. ab140607; Abcam) and anti-CD163 (1:500, ab182422; Abcam) antibodies overnight at 4°C as previously reported (24). For statistical analysis, the percentage coverage of the protein was scored manually as follows: i) 1 (0–25%); ii) 2 (26–50%); iii) 3 (51–75%); and iv) 4 (76–100%) (24). The intensity of positive staining was also scored as follows: i) 0 (negative); ii) 1 (weak); iii) 2 (moderate); and iv) 3 (strong). The final scores were calculated by multiplying the aforementioned scores. The final scores of the percentage and staining scores were defined as the overall IHC scores (0–12). Scores <6 were considered to be low expression (STX6-Low) and scores ≥6 were considered to be high expression (STX6-High).
Gene expression profiling interactive analysis (GEPIA2) database analysis
GEPIA2 (http://gepia2.cancer-pku.cn/#index) is an interactive online platform, which contains information from >9,000 tumor samples from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) and The Genotype-Tissue Expression (GTEx) (https://xenabrowser.net/datapages/) databases, and information from >8,000 control samples. The correlation between STX6 and CD163 was determined by Pearson correlation coefficient analysis in GEPIA2 (25). The iCluster 1 and iCluster 3 datasets (Cutoff-High-75% and Cutoff-Low-25%) were used to analyze the association between prognosis and STX6 expression in patients with HCC (25).
Kaplan-Meier plotter (KM plotter) survival analysis of STX6
Kaplan-Meier plotter (http://kmplot.com/analysis/) can perform survival analysis on >54,000 genes (mRNA, microRNA and protein) in 21 types of tumors (including breast, ovarian, lung, gastric and liver cancer). The data mainly come from the Gene Expression Omnibus and TCGA databases (26). The results were assessed based on the log rank P-value and hazard ratio (HR) with 95% confidence intervals. The RNAseq ID: 10228 (STX6) and Cutoff value used in analysis: 1046 were used. A follow-up threshold of 60 months was used to analyze the association between prognosis and STX6 expression in patients with HCC based on the log rank P-value.
Tumor immune estimation resource (TIMER) database
The TIMER database (https://cistrome.shinyapps.io/timer/) (27) is mainly divided into seven sections: Gene, Survival, Mutation, somatic copy number amplifications (sCNA), differential expression (Diffexp), Correlation and Estimation. Among them, the Gene and Correlation were used in this study. To facilitate the study of tumor immunity and genomic data, the TIMER database applies a deconvolution method (28) to infer the abundance of tumor immune-infiltrating cells (TIICs) from gene expression profiles, reanalyzes the gene expression data of 10,897 samples of 32 cancer types from TCGA and estimates the abundance of 6 TIIC subgroups [B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils and dendritic cells (DCs)]. Deconvolution methods define the problem as mathematical equations that model the gene expression of a tissue sample as the weighted sum of the expression profiles from the cells in the population mix (29). These methods are further detailed in previous studies (27,30). The statistical method used by the TIMER database in the present study was the Spearman correlation coefficient analysis. P<0.05 was considered to be statistically significant.
University of California Santa Cruz (UCSC) Xena
RNAseq data were extracted from the UCSC Xena portal (https://xenabrowser.net/datapages/) (31), including data from TCGA Liver HCC (LIHC; n=371) and GTEx for corresponding normal tissue (n=160). These data were used to assess the diagnostic effect of STX6 and α-fetoprotein (AFP) in liver cancer using a receiver operating characteristic (ROC) curve as previously reported (32).
Statistical analysis
All bioinformatics analyses were performed using the corresponding database websites. Other statistical analyses of data were performed using GraphPad Prism 6 (GraphPad Software, Inc.) and SPSS 18 (SPSS, Inc.). Data are presented as the median. The Wilcoxon signed-rank test was used to compare the difference between tumor tissues and normal tissues. A Mann-Whitney U test was used to compare two distinct groups of patients (STX6-Low vs. STX6-High). The association of clinicopathological factors with STX6 expression was analyzed using the χ2 test. A log-rank test was used to compare the survival curves by Kaplan-Meier. The correlation between the IHC score of STX6 and the IHC score of CD163 was analyzed using Pearson correlation coefficient analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
STX6 is highly expressed in HCC
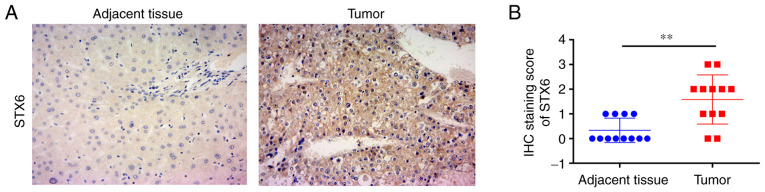
IHC was used to assess the protein expression levels of STX6 in patients with HCC (Fig. 1A). The results demonstrated that the IHC scores of 12 pairs of HCC samples stained for STX6 were significantly higher than those of paired adjacent tissues (Fig. 1B). These results demonstrated that STX6 protein was highly expressed in HCC tissues.
Figure 1.
STX6 protein expression in human HCC and paired adjacent tissues. (A) IHC staining of STX6 in HCC and paired adjacent tissues (magnification, ×100). (B) STX6 protein expression presented as the IHC staining score in 12 HCC and paired adjacent tissues. Data are presented as the mean ± SD. **P<0.01. HCC, hepatocellular carcinoma; IHC, immunohistochemistry; STX6, syntaxin 6.
Association analysis between STX6 expression and clinical features of patients with HCC
The relationship between STX6 protein expression and the clinicopathological characteristics of 90 patients with HCC was evaluated. STX6 protein expression was significantly associated with HCC tumor size (P=0.003), Edmondson grade (P=0.020) and AFP level (P=0.019) (Table I).
Table I.
Association between STX6 expression and clinical characteristics of patients with hepatocellular carcinoma.
| STX6 expression (IHC score) | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Variables | No. (%) | Low (<6), n | High (≥6), n | Df | χ2 | P-value |
| Sex | 1 | 0.017 | 0.897 | |||
| Male | 76 (84.4) | 42 | 34 | |||
| Female | 14 (15.6) | 8 | 6 | |||
| Age, years | 1 | 0.057 | 0.810 | |||
| <55 | 37 (41.1) | 20 | 17 | |||
| ≥55 | 53 (58.9) | 30 | 23 | |||
| Size, cm | 1 | 9.085 | 0.003 | |||
| <5 | 56 (62.2) | 38 | 18 | |||
| ≥5 | 34 (37.8) | 12 | 22 | |||
| Diolame complete | 1 | 0.188 | ||||
| Yes | 54 | 31 | 23 | 0.665 | ||
| No | 36 | 19 | 17 | |||
| Number of tumors | 1 | 1.125 | 0.289 | |||
| Single | 72 (80) | 38 | 34 | |||
| Multiple | 18 (20) | 12 | 6 | |||
| TNM staging | 1 | 1.798 | 0.180 | |||
| I+II | 77 (85.6) | 45 | 32 | |||
| III+IV | 13 (14.4) | 5 | 8 | |||
| Microvascular invasion | 1 | 1.309 | 0.253 | |||
| No | 64 (71.1) | 38 | 26 | |||
| Yes | 26 (28.9) | 12 | 14 | |||
| Edmondson grade | 1 | 5.399 | 0.020 | |||
| I+II | 67 (74.4) | 42 | 25 | |||
| III | 23 (25.6) | 8 | 15 | |||
| Cirrhosis | 1 | 0.243 | 0.622 | |||
| Negative | 16 (17.8) | 8 | 8 | |||
| Positive | 74 (82.2) | 42 | 32 | |||
| HBV | 1 | 0.800 | 0.371 | |||
| Absent | 25 (27.8) | 12 | 13 | |||
| Present | 65 (72.2) | 38 | 27 | |||
| ALT, U/l | 1 | 0.458 | 0.499 | |||
| <45 | 55 (61.1) | 29 | 26 | |||
| ≥45 | 35 (38.9) | 21 | 14 | |||
| AFP, ng/ml | 1 | 5.478 | 0.019 | |||
| <400 | 69 (76.7) | 43 | 26 | |||
| ≥400 | 21 (23.3) | 7 | 14 | |||
AFP, α-fetoprotein; ALT, alanine aminotransferase; HBV, hepatitis B virus; IHC, immunohistochemistry; STX6, syntaxin 6.
Association between STX6 expression and patient survival in HCC
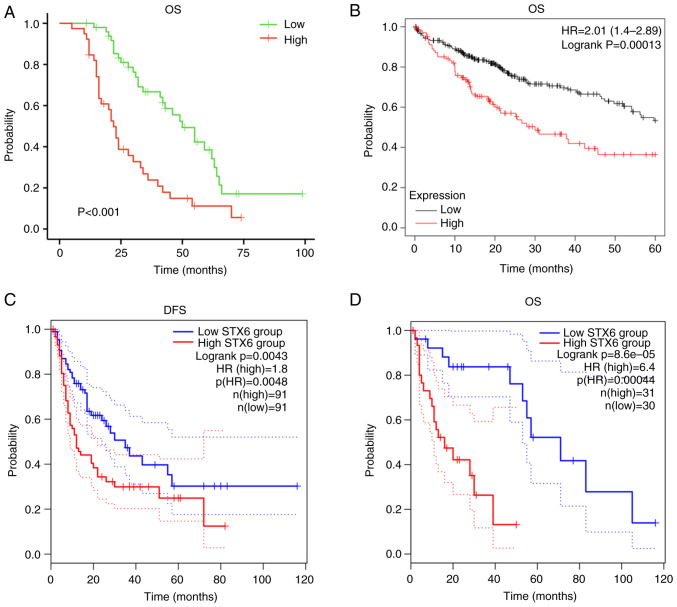
Survival analysis demonstrated that patients with high STX6 expression had worse prognosis (Fig. 2A). Furthermore, analysis using KM plotter (Fig. 2B) demonstrated that high expression levels of STX6 were associated with worse overall survival (OS; P=0.00013; HR, 2.01; 95% CI, 1.4-2.89) in patients with HCC. Additionally, in the GEPIA2 database analysis, high STX6 expression was associated with worse disease-free survival [P=0.0043; HR(high), 1.8; p(HR), 0.0048] and OS [P=0.000086; HR, 6.4; p(HR)=0.00044] in patients with HCC (Fig. 2C and D). These data demonstrated that dysregulated expression of STX6 affected the clinical outcomes of patients with HCC.
Figure 2.
Predictive value of STX6 for prognosis in HCC. (A) OS in patients with the present HCC data. (B) OS analyzed using Kaplan-Meier plotter. (C) DFS and (D) OS analyzed using Gene Expression Profiling Integrative Analysis 2. DFS, disease-free survival; HCC, hepatocellular carcinoma; HR, hazard ratio; OS, overall survival; STX6, syntaxin 6.
Analysis of immune infiltration
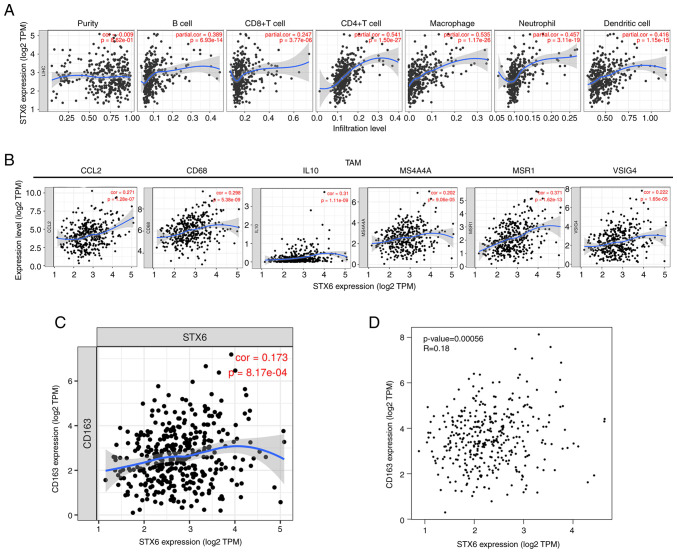
STX6 expression was significantly positively correlated with infiltration by B cells (r=0.389; P=6.93×10−14), CD4+ T cells (r=0.541; P=1.50×10−27), macrophages (r=0.535; P=1.17×10−26), neutrophils (r=0.457; P=3.11×10−19) and DCs (r=0.416; P=1.15×10−15), and significantly positively associated with CD8+ T cell infiltration (r=0.247; P=3.77×10−6; Fig. 3A), which demonstrated that STX6 serves a crucial role in the immune infiltration of HCC. STX6 expression was also significantly positively associated with macrophage TAMs: CCL2 (r=0.271; P=1.28×10−7), CD68 (r=0.298; P=5.38×10−9), IL10 (r=0.31; P=1.11×10−9), MS4A4A (r=0.202; P=9.06×10−5), MSR1 (r=0.371; P=1.62×10−13) and VSIG4 (r=0.222; P=1.65×10−5) were also analyzed (Fig. 3B). The results demonstrated that STX6 mRNA expression was significantly correlated with TAMs [CC motif chemokine ligand 2 (CCL2), CD68, IL10, V-set and immunoglobulin domain containing 4, macrophage scavenger receptors-type 1 and membrane spanning 4-domains A4A], DCs, CD4+ T cells, CD163, etc. (Table SI). To further assess the association of STX6 with CD163, STX6 and CD163 expression was analyzed using the GEPIA2 and TIMER databases. These results demonstrated a small association between STX6 and CD163 (cor=0.173 in Fig. 3C and R=0.18 in Fig. 3D). The results of the present study demonstrated that STX6 expression was positively associated with B and T-cell receptor signaling pathways during HCC pathogenesis, indicating that STX6 was related to the immune response. These findings suggested that STX6 expression may be associated with the infiltration of immune cells in HCC.
Figure 3.
Relationship between STX6 expression and immune cell infiltration levels in hepatocellular carcinoma. (A) Correlation of STX6 expression with tumor-infiltrating immune cells in LIHC (n=371). Scatter plots presenting the correlations between STX6 expression and marker molecules, including (B) TAMs (CCL2, CD68, IL10, VSIG4, MSR1, MS4A4A), and (C and D) CD163. The blue curve and gray area in the figure represent the general trend direction. LIHC, liver hepatocellular carcinoma; STX6, syntaxin 6; CCL2, CC motif chemokine ligand 2; MS4A4A, membrane spanning 4-domains A4A; MSR1, macrophage scavenger receptor 1; TAMs, tumor-associated macrophages; TPM, transcripts per million; VSIG4, V-set and immunoglobulin domain containing 4.
STX6 is positively associated with CD163 according to IHC
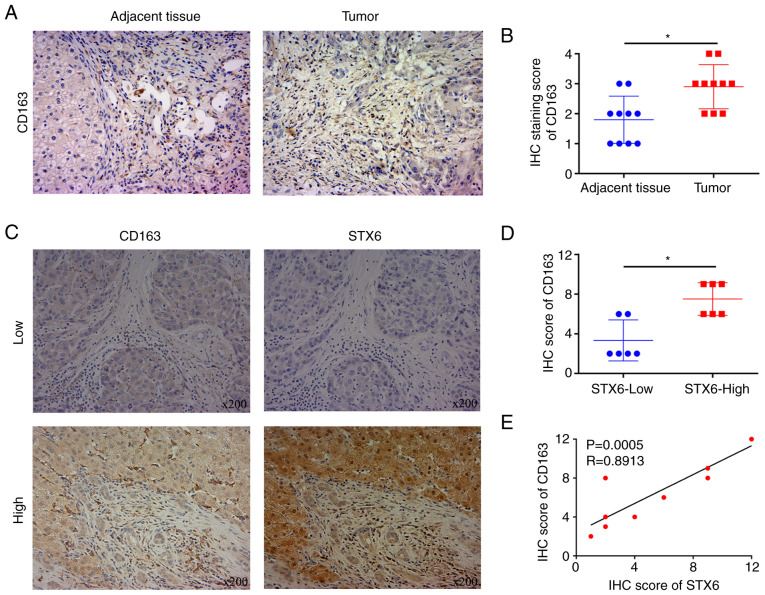
STX6 and CD163 expression in HCC and adjacent noncancerous tissues was assessed using IHC (Figs. 1A and 4A). The results demonstrated that the protein expression levels of STX6 and CD163 in cancer tissues were significantly increased compared with those in adjacent tissues (Figs. 1B and 4B). To further examine the association between STX6 and CD163, IHC was performed (Fig. 4C). The results demonstrated that CD163 levels were significantly positively associated with the levels of STX6 in paired paraffin-embedded tumor slices (Fig. 4D and E).
Figure 4.
Expression levels of STX6 and CD163 in HCC. (A) CD163 expression in HCC and adjacent noncancerous tissues (magnification, ×100). (B) IHC staining score of CD163 in HCC and adjacent noncancerous tissues. (C) STX6 and CD163 expression in paired paraffin-embedded tumor slices (magnification, ×100). (D) IHC score of CD163 in tumor tissues from patients in the high and low STX6 groups. (E) Correlation between the IHC score of STX6 and the IHC score of CD163. *P<0.05. HCC, hepatocellular carcinoma; IHC, immunohistochemistry; STX6, syntaxin 6.
Diagnostic values of STX6 and AFP as assessed using ROC curve analyses
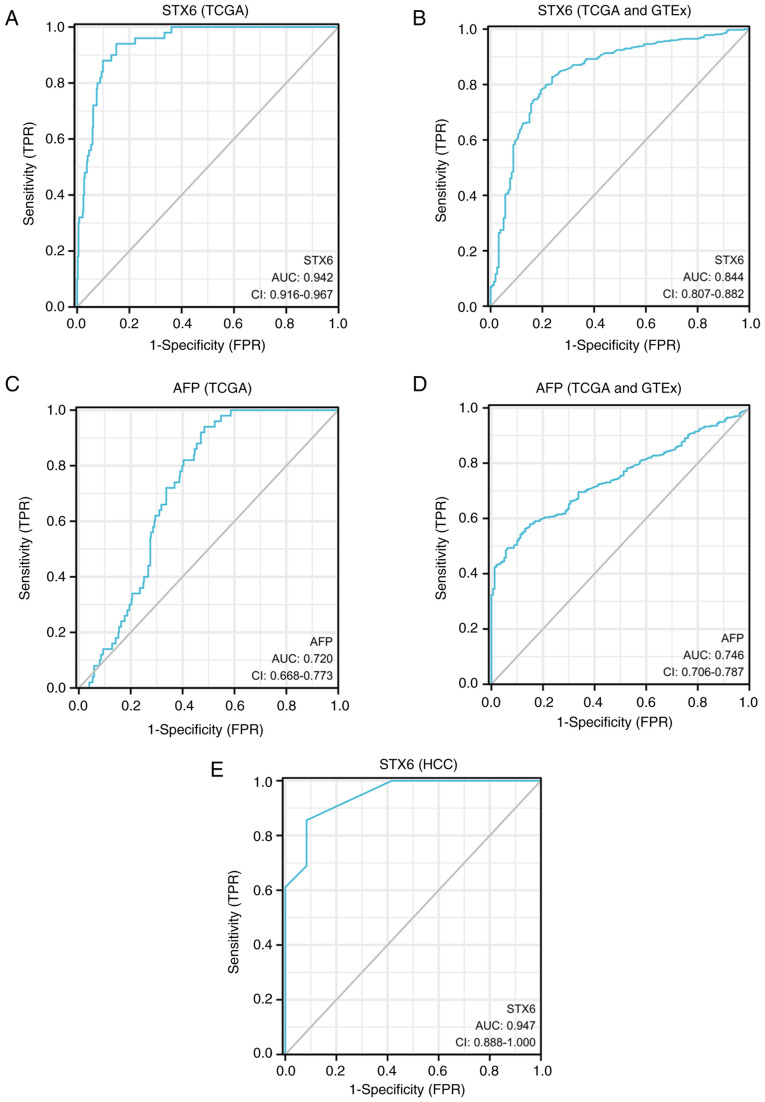
The ROC curve analyses of two markers (STX6 and AFP) in HCC and normal tissues are presented (Fig. 5). ROC curve analysis indicated that the area under the curve (AUC) values for STX6 and AFP were 0.942 (CI, 0.916-0.967) and 0.720 (CI, 0.668-0.773), respectively, using TCGA data (tumor, n=374; normal, n=50). AUC of STX6 (sensitivity, 0.940; specificity, 0.850) was higher than that of AFP (sensitivity, 0.940; specificity, 0.516) in TCGA (Table II). Furthermore, the ROC curve analysis demonstrated that the AUC values of STX6 and AFP were 0.844 and 0.746, respectively, according to the combined TCGA (tumor, n=371) and GTEx (normal, n=160) datasets. The sensitivity of the ROC curve for STX6 (0.827) was higher than that for AFP (0.563). The specificity of the ROC curve of STX6 (0.762) was lower than that of AFP (0.869). Compared with the aforementioned datasets, STX6 had a higher AUC value (0.947) and specificity (0.917) in the our HCC data of the present study. The sensitivity of ROC curve of STX6 is 0.856 in the present HCC data. These results demonstrated that STX6 may be a diagnostic marker for HCC.
Figure 5.
Diagnostic values of STX6 and AFP indicated by ROC curve analyses based on the TCGA and GTEx datasets. (A) Diagnostic value of STX6 assessed using ROC curve analysis of data from TCGA (tumor, n=374; normal, n=50). (B) Diagnostic value of STX6 assessed using ROC curve analysis of data from TCGA (tumor, n=371) and GTEx (normal, n=160) databases. (C) Diagnostic value of AFP assessed using ROC curve analysis of data from TCGA (tumor, n=374; normal, n=50). (D) Diagnostic value of AFP assessed using ROC curve analysis of data from TCGA (tumor, n=371) and GTEx (normal, n=160) databases. (E) Diagnostic value of STX6 assessed using ROC curve analysis of data from the present study (tumor, n=90; normal, n=12). AFP, α-fetoprotein; AUC, area under the curve; FPR, false positive rate; GTEx, Genotype-Tissue Expression; HCC, hepatocellular carcinoma; ROC, receiver operating characteristic; STX6, syntaxin 6; TCGA, The Cancer Genome Atlas; TPR, true positive rate.
Table II.
Diagnostic values of STX6 and AFP in patients with liver hepatocarcinoma according to receiver operating characteristic curve analyses.
| Index | AUC | 95% CI | Cut-off | Sensitivity | Specificity | Youden index | Data |
|---|---|---|---|---|---|---|---|
| AFP | 0.720 | 0.668-0.773 | 2.613 | 0.940 | 0.516 | 1.456 | TCGA |
| STX6 | 0.942 | 0.916-0.967 | 2.334 | 0.940 | 0.850 | 1.790 | TCGA |
| AFP | 0.746 | 0.706-0.787 | 3.341 | 0.563 | 0.869 | 1.432 | TCGA + GTEx |
| STX6 | 0.844 | 0.807-0.882 | 1.702 | 0.827 | 0.762 | 1.590 | TCGA + GTEx |
| STX6 | 0.947 | 0.888-1.000 | 1.500 | 0.856 | 0.917 | 0.772 | HCC |
TCGA data (tumor, n=374; normal, n=50). TCGA (Tumor=371) + GTEx (normal=160) data. HCC data (tumor, n=90; normal, n=12). AFP, α-fetoprotein; AUC, area under the curve; CI, confidence interval; GTEx, Genotype-Tissue Expression; HCC, hepatocellular carcinoma; STX6, syntaxin 6; TCGA, The Cancer Genome Atlas.
Discussion
The progression of liver HCC is rapid and numerous patients present with advanced HCC (33,34). Regulation of immune infiltration is increasingly recognized as being important in tumor development (35). Finding breakthroughs in immunotherapy has become the focus of current research. Furthermore, an effective early detection method is still lacking in the current treatment of HCC. AFP has been recognized as a tumor marker for HCC but it has poor sensitivity and specificity (36,37). Exploring novel therapeutic and diagnostic markers is still the top priority of scientific research in this field.
In the present study, the role of the STX6 gene in the development of HCC was analyzed using IHC. STX6 protein expression in HCC was analyzed and the results demonstrated that STX6 expression was upregulated in HCC tissues compared with normal tissues. The association between STX6 and clinical characteristics of patients was further considered. These results demonstrated that high protein expression levels of STX6 were significantly associated with tumor size, Edmondson grade and the AFP level in patients with HCC. Furthermore, these results demonstrated that patients with high STX6 expression had a worse prognosis as demonstrated by analysis of the survival data in the KM plotter and GEPIA2 databases, which are based on the results of transcriptome sequencing data analysis. Furthermore, patients with high STX6 protein expression also had a worse prognosis as demonstrated by the survival data of patients with HCC in the present study. A previous study reported that STX6 could be a prognostic biomarker for patients with renal cell carcinoma based on TCGA transcriptome sequencing data (9). This is supported by the present study which combined analysis of two different datasets, which demonstrated that STX6 could promote the process of HCC malignancy.
The present study demonstrated that STX6 was associated with infiltration of immune cells based on analysis using the TIMER database. The results demonstrated that high STX6 mRNA expression in HCC was positively associated with high immune infiltration. STX6 expression was significantly positively correlated with the levels of immune infiltration, including B cell, CD4+ T cell, DC, macrophage and neutrophil infiltration, in HCC, and significantly positively associated with CD8+ T cells. Furthermore, the gene markers of M2 macrophages (CD163 and CD115) and TAMs (CCL2 and IL10) were significantly positively correlated with STX6 expression. Association between STX6 and CD163 was assessed using IHC and the results confirmed the aforementioned findings. It could be concluded that the M2 macrophage CD163 expression was also enhanced by increased levels of STX6. It has previously been reported that high rates of infiltration of M2 macrophages into the tumor stroma could inhibit T cell proliferation and downregulate antitumor immune responses (38,39). STX6 upregulation could be one of the routes that link immunosuppression and the development of HCC. Overall, these results demonstrated the potential regulatory role of STX6 in immune inflammatory responses in HCC.
AFP has a diagnostic value as a marker for liver cancer in clinical settings (40–42). Abnormal AFP levels in adult plasma have been reported to be a marker of the pathological condition of HCC (43). In the present study, the results demonstrated that the AUC of STX6 was significantly higher than the AUC of AFP based on TCGA data (0.942 vs. 0.720) and the sensitivity of STX6 was also higher than that of AFP based on combined TCGA and GTEx data (0.827 vs. 0.563). Nevertheless, the specificity of the AUC curve of STX6 was lower than the AUC of AFP based on combined TCGA (tumor, n=371) and GTEx (normal, n=160) data (0.762 vs. 0.869). This could have been due to the high STX6 expression in multiple other tumor types (7,8,44), which reduce its diagnostic specificity in HCC. However, compared with the aforementioned datasets, STX6 demonstrated a higher AUC value (0.947) and specificity (0.917) in data from the present study. Collectively, these results demonstrated that STX6 has the potential to be a powerful diagnostic maker in HCC.
The present study demonstrated that STX6 protein expression was significantly associated with HCC tumor size, Edmondson grade, AFP level and prognosis of patients with HCC. Furthermore, STX6 may be involved in the immune-inflammatory response of HCC and may become a novel potential diagnostic marker for patients with HCC. However, there were some limitations in the present study. For the assessment of the association between STX6 and CD163, the absence of double-staining IHC or immunofluorescence staining was a limitation of the present study. Furthermore, the present study was only an initial early-stage experiment and future work is required which should include experiments on larger numbers of clinical samples and more in-depth research on the molecular mechanism of STX6 in HCC. Further studies should analyze fresh tissues and HCC cell lines and explore the effects of STX6 on the phenotype of HCC cells.
In summary, the present study demonstrated that STX6 expression was associated with the clinical characteristics and prognosis of patients with HCC. Furthermore, STX6 may associate with CD163 to participate in the modulation of inflammatory responses in HCC. Compared with AFP, STX6 may become a valuable novel tumor marker for the diagnosis of patients with HCC and combination of STX6 with AFP may have higher diagnostic value. The present study also provided further insight into the molecular mechanism of STX6 in HCC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant no. 82103165), The Key R & D General Project of Jiangxi Science and Technology Department (grant no. 20203BBGL73187), the Natural Science Foundation of Jiangxi Province (grant no. 20212BAB216036), The Education Department of Jiangxi Province Science and Technology Research Projects (grant no. GJJ160246) and the Young Talent Cultivation Project of the First Affiliated Hospital of Nanchang University (grant no. YFYPY202007).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the GEPIA2 repository, https://gepia2.cancer-pku.cn/#index; National Cancer Institute GDC Data Portal repository, https://portal.gdc.cancer.gov/; the Kaplan-Meier Plotter repository, https://kmplot.com/analysis/; TIMER repository, https://cistrome.shinyapps.io/timer/; TCGA repository, https://portal.gdc.cancer.gov/; and UCSC Xena repository [TCGA cohort, GDC TCGA Liver Cancer (LIHC), TCGA-LIHC.htseq_fpkm.tsv; GTEX cohort, GTEX, gtex_RSEM_gene_tpm], https://xenabrowser.net/datapages/. The other datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YZ, LL, ZF and YT analyzed the data and prepared the manuscript. YL and JX designed the study. YL and JX confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The paired paraffin-embedded tumor slices were obtained from Human Genetic Resources Center of the First Affiliated Hospital of Nanchang University (Nanchang, China). The project was approved by the Clinical Medical Research Ethics Committee of the First Affiliated Hospital of Nanchang University (approval no. 202112020; Nanchang, China). As this was a retrospective study, the requirement for informed consent was waived by the ethics committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873:188314. doi: 10.1016/j.bbcan.2019.188314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orcutt ST, Anaya DA. Liver resection and surgical strategies for management of primary liver cancer. Cancer Control. 2018;25:1073274817744621. doi: 10.1177/1073274817744621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Höglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, Rademakers R, de Silva R, Litvan I, Riley DE, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- 5.Bock JB, Lin RC, Scheller RH. A new syntaxin family member implicated in targeting of intracellular transport vesicles. J Biol Chem. 1996;271:17961–17965. doi: 10.1074/jbc.271.30.17961. [DOI] [PubMed] [Google Scholar]
- 6.Wendler F, Tooze S. Syntaxin 6: The promiscuous behaviour of a SNARE protein. Traffic. 2001;2:606–611. doi: 10.1034/j.1600-0854.2001.20903.x. [DOI] [PubMed] [Google Scholar]
- 7.Du J, Liu X, Wu Y, Zhu J, Tang Y. Essential role of STX6 in esophageal squamous cell carcinoma growth and migration. Biochem Biophys Res Commun. 2016;472:60–67. doi: 10.1016/j.bbrc.2016.02.061. [DOI] [PubMed] [Google Scholar]
- 8.Zhang PR, Ren J, Wan JS, Sun R, Li Y. Circular RNA hsa_circ_0002052 promotes osteosarcoma via modulating miR-382/STX6 axis. Hum Cell. 2020;33:810–818. doi: 10.1007/s13577-020-00335-9. [DOI] [PubMed] [Google Scholar]
- 9.Peak TC, Su Y, Chapple AG, Chyr J, Deep G. Syntaxin 6: A novel predictive and prognostic biomarker in papillary renal cell carcinoma. Sci Rep. 2019;9:3146. doi: 10.1038/s41598-019-39305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West ZE, Aitcheson SM, Semmler ABT, Murray RZ. The trans-SNARE complex VAMP4/Stx6/Stx7/Vti1b is a key regulator of Golgi to late endosome MT1-MMP transport in macrophages. Traffic. 2021;22:368–376. doi: 10.1111/tra.12813. [DOI] [PubMed] [Google Scholar]
- 11.Bian Z, Shi L, Kidder K, Zen K, Garnett-Benson C, Liu Y. Intratumoral SIRPα-deficient macrophages activate tumor antigen-specific cytotoxic T cells under radiotherapy. Nat Commun. 2021;12:3229. doi: 10.1038/s41467-021-23442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/S1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi T, Fushida S, Yamamoto Y, Tsukada T, Kinoshita J, Oyama K, Miyashita T, Tajima H, Ninomiya I, Munesue S, et al. Tumor-associated macrophages of the M2 phenotype contribute to progression in gastric cancer with peritoneal dissemination. Gastric Cancer. 2016;19:1052–1065. doi: 10.1007/s10120-015-0579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munir MT, Kay MK, Kang MH, Rahman MM, Al-Harrasi A, Choudhury M, Moustaid-Moussa N, Hussain F, Rahman SM. Tumor-associated macrophages as multifaceted regulators of breast tumor growth. Int J Mol Sci. 2021;22:6526. doi: 10.3390/ijms22126526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cersosimo F, Lonardi S, Bernardini G, Telfer B, Mandelli GE, Santucci A, Vermi W, Giurisato E. Tumor-associated macrophages in osteosarcoma: From mechanisms to therapy. Int J Mol Sci. 2020;21:5207. doi: 10.3390/ijms21155207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, Liu Q, Dou R, Xiong B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18:64. doi: 10.1186/s12943-019-0976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arvanitakis K, Koletsa T, Mitroulis I, Germanidis G. Tumor-associated macrophages in hepatocellular carcinoma pathogenesis, prognosis and therapy. Cancers (Basel) 2022;14:226. doi: 10.3390/cancers14010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 19.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawamura K, Komohara Y, Takaishi K, Katabuchi H, Takeya M. Detection of M2 macrophages and colony-stimulating factor 1 expression in serous and mucinous ovarian epithelial tumors. Pathol Int. 2009;59:300–305. doi: 10.1111/j.1440-1827.2009.02369.x. [DOI] [PubMed] [Google Scholar]
- 21.Bao D, Zhao J, Zhou X, Yang Q, Chen Y, Zhu J, Yuan P, Yang J, Qin T, Wan S, Xing J. Mitochondrial fission-induced mtDNA stress promotes tumor-associated macrophage infiltration and HCC progression. Oncogene. 2019;38:5007–5020. doi: 10.1038/s41388-019-0772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bover LC, Cardó-Vila M, Kuniyasu A, Sun J, Rangel R, Takeya M, Aggarwal BB, Arap W, Pasqualini R. A previously unrecognized protein-protein interaction between TWEAK and CD163: Potential biological implications. J Immunol. 2007;178:8183–8194. doi: 10.4049/jimmunol.178.12.8183. [DOI] [PubMed] [Google Scholar]
- 23.Murray RZ, Wylie FG, Khromykh T, Hume DA, Stow JL. Syntaxin 6 and Vti1b form a novel SNARE complex, which is up-regulated in activated macrophages to facilitate exocytosis of tumor necrosis factor-alpha. J Biol Chem. 2005;280:10478–10483. doi: 10.1074/jbc.M414420200. [DOI] [PubMed] [Google Scholar]
- 24.Xiong J, Feng Z, Li Z, Zhong T, Yang Z, Tu Y, Xiao T, Jie Z, Cao Y. Overexpression of TWA1 predicts poor prognosis in patients with gastric cancer. Pathol Res Pract. 2019;215:152594. doi: 10.1016/j.prp.2019.152594. [DOI] [PubMed] [Google Scholar]
- 25.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45((W1)):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lánczky A, Nagy Á, Bottai G, Munkácsy G, Szabó A, Santarpia L, Győrffy B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/1538-7445.AM2017-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman C, Fridman WH, de Reyniès A. Erratum to: Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:249. doi: 10.1186/s13059-016-1113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. 2017;6:e26476. doi: 10.7554/eLife.26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48((W1)):W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vivian J, Rao AA, Nothaft FA, Ketchum C, Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD, Musselman-Brown A, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35:314–316. doi: 10.1038/nbt.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34:153–159. doi: 10.1053/j.semdp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156:477–491.e1. doi: 10.1053/j.gastro.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan Y, Yu Y, Wang X, Zhang T. Tumor-associated macrophages in tumor immunity. Front Immunol. 2020;11:583084. doi: 10.3389/fimmu.2020.583084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Debruyne EN, Delanghe JR. Diagnosing and monitoring hepatocellular carcinoma with alpha-fetoprotein: New aspects and applications. Clin Chim Acta. 2008;395:19–26. doi: 10.1016/j.cca.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Lu Z, Zuo B, Jing R, Gao X, Rao Q, Liu Z, Qi H, Guo H, Yin H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J Hepatol. 2017;67:739–748. doi: 10.1016/j.jhep.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 38.Lee WC, Reuben A, Hu X, McGranahan N, Chen R, Jalali A, Negrao MV, Hubert SM, Tang C, Wu CC, et al. Multiomics profiling of primary lung cancers and distant metastases reveals immunosuppression as a common characteristic of tumor cells with metastatic plasticity. Genome Biol. 2020;21:271. doi: 10.1186/s13059-020-02175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho DW, Tsui YM, Chan LK, Sze KM, Zhang X, Cheu JW, Chiu YT, Lee JM, Chan AC, Cheung ET, et al. Single-cell RNA sequencing shows the immunosuppressive landscape and tumor heterogeneity of HBV-associated hepatocellular carcinoma. Nat Commun. 2021;12:3684. doi: 10.1038/s41467-021-24010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galle PR, Foerster F, Kudo M, Chan SL, Llovet JM, Qin S, Schelman WR, Chintharlapalli S, Abada PB, Sherman M, Zhu AX. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019;39:2214–2229. doi: 10.1111/liv.14223. [DOI] [PubMed] [Google Scholar]
- 41.Luo P, Wu S, Yu Y, Ming X, Li S, Zuo X, Tu J. Current status and perspective biomarkers in AFP negative HCC: Towards screening for and diagnosing hepatocellular carcinoma at an earlier stage. Pathol Oncol Res. 2020;26:599–603. doi: 10.1007/s12253-019-00585-5. [DOI] [PubMed] [Google Scholar]
- 42.Kim KI, Chung HK, Park JH, Lee YJ, Kang JH. Alpha-fetoprotein-targeted reporter gene expression imaging in hepatocellular carcinoma. World J Gastroenterol. 2016;22:6127–6134. doi: 10.3748/wjg.v22.i27.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen T, Dai X, Dai J, Ding C, Zhang Z, Lin Z, Hu J, Lu M, Wang Z, Qi Y, et al. AFP promotes HCC progression by suppressing the HuR-mediated Fas/FADD apoptotic pathway. Cell Death Dis. 2020;11:822. doi: 10.1038/s41419-020-03030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Y, Ye Z, Lu G, Yang N, Zhang J, Wang L, Cui J, Del Oozo MA, Wu Y, Xia D, Shen HM. Cholesterol-enriched membrane micro-domaindeficiency induces doxorubicin resistancevia promoting autophagy in breast cancer. Mol Ther Oncolytics. 2021;23:311–329. doi: 10.1016/j.omto.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the GEPIA2 repository, https://gepia2.cancer-pku.cn/#index; National Cancer Institute GDC Data Portal repository, https://portal.gdc.cancer.gov/; the Kaplan-Meier Plotter repository, https://kmplot.com/analysis/; TIMER repository, https://cistrome.shinyapps.io/timer/; TCGA repository, https://portal.gdc.cancer.gov/; and UCSC Xena repository [TCGA cohort, GDC TCGA Liver Cancer (LIHC), TCGA-LIHC.htseq_fpkm.tsv; GTEX cohort, GTEX, gtex_RSEM_gene_tpm], https://xenabrowser.net/datapages/. The other datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.