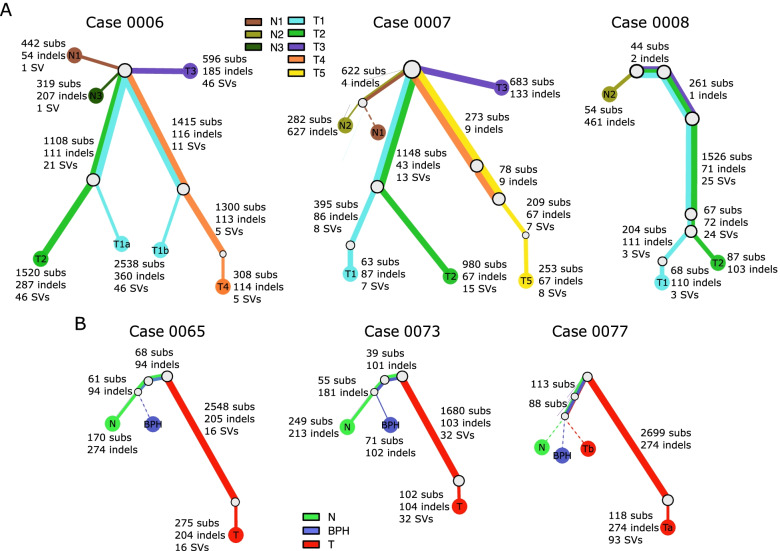
Fig. 3.
Phylogenies of patients with multiple samples. Phylogenies revealing the relationships between clones for each case. A patients where we have collected multiple tumours and normal. B patients where there was data from a tumour, non-BPH normal tissue, and BPH normal tissue. Each coloured line represents a clone/subclone detected in a particular sample. When two or more coloured lines are together, they represent a clone that is found in all the samples represented. The length of the line is proportional to the weighted number of single nucleotide variants present in each clone; the thickness represents the clonal cell fraction associated with that clone (more detail in Additional file 3). For example, case 0077 contains a shared subclone with 8% N, 33% BPH and 2% T (Tb) supported by 113 SNVs and 4 indels. Dotted lines are associated with samples that have no evidence of a unique sample specific clone. The very low fraction tumour subclone (< 4%) shared with normal and BPH tissue in case 0077 and between normal and tumour in case 0072 suggests cancer targeted tissue contained some of the N/BPH cells. Additional phylogenies can be found in Supplementary Fig. 3