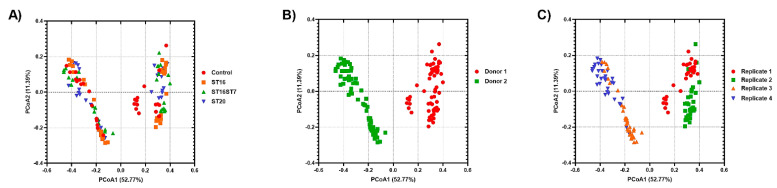
Figure 3.
Plots of principal coordinate analysis (PCoA) based on Bray–Curtis distances among the identified microbiota in different samples, showing clustering based on treatment (A); donor (B), and replicate (C). The samples were colored as indicated in the legends. PCoA1 and PCoA2 represent the top two coordinates that captured the highest microbial variability among samples, and the percentage shown indicates the fraction of variation represented by each coordinate. Permutational multivariate analysis of variance (PERMANOVA) was used to test for the statistical significance of sample grouping.