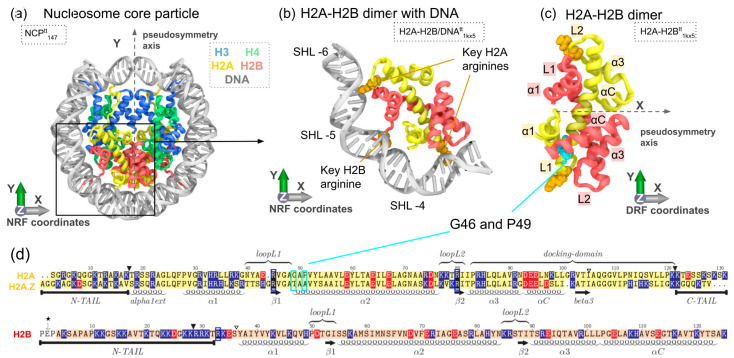
Figure 1.
Overview of the nucleosome core particle, H2A-H2B dimer structure, their elements and analyzed systems. (a) Nucleosome core particle (NCP). (b) H2A-H2B dimer. (c) H2A-H2B dimer bound to 30 DNA base pairs, H2A key arginines, which interact with the DNA minor groove on both sides of the H2A α2-helix, are shown in orange. Introduced reference frames (NRF—nucleosome reference frame, DRF—dimer reference frame) are shown on each panel. (d) H2B histone sequence and H2A-H2A.Z sequence alignment (Xenopus laevis canonical H2A and Homo sapiens H2A.Z). Basic and acidic residues are colored blue and red, respectively. Anchor arginines are highlighted by blue frames. Key structural elements are annotated below or on top of the sequences. Two positions on the alignment (highlighted in cyan frames) correspond to analog positions of H2A/H2A.Z substitutions in yeast affecting nucleosome stability and SWR1 activity [39,40]. ▼ and ▽ mark the histone tails’ truncation sites in NCP and dimer systems’ simulations, respectively.