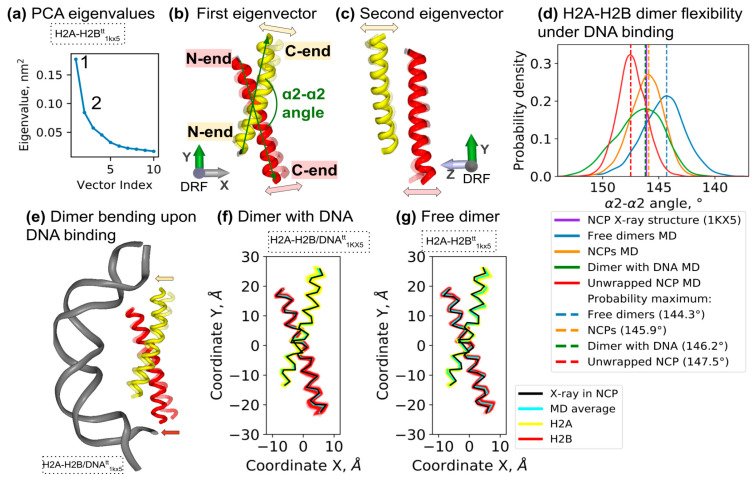
Figure 3.
Intrinsic H2A-H2B dimer bending and effects of interaction with DNA. (a–c) Principal component analysis of histone α2-helices’ Cα-atoms thermal fluctuations in MD. (a) Eigenvalues of the first 10 eigenvectors. (b,c) Dynamical modes corresponding to first and second eigenvectors. Snapshots of two extreme states along the eigenvectors are shown (see Figure S6 and interactive materials for a more detailed view). The angle between the two α2-helices is shown in (b) (see Methods for definitions). (d) Probability distributions of α2-α2 angle values during MD simulations of different systems: free H2A-H2B dimers (systems H2A-H2Btt1KX5 and H2A-H2Btt3LZ0), dimer bound to DNA (H2A-H2B/DNAtt1KX5) and NCPs (averaged from NCP147, NCPtt147, NCPtt146, NCPtt145). The dashed vertical lines mark the maximum distributions, and the solid violet vertical line marks the value in 1KX5 X-ray NCP structure. The corresponding plots for all simulated systems are shown in Figure S8. For statistical analysis, see Table S3. (e) Schematic representation of H2A-H2B bending upon DNA binding. Arrows mark the direction of the bending. (f,g) Two-dimensional projections of histone α2-helices conformations in MD of free (f) and DNA-bound dimer (g). The mean helix conformations and conformations in an NCP X-ray structure are shown.