Abstract
Simple Summary
Gut microbial dysbiosis and microbial passage into the peripheral blood leads to colorectal cancer (CRC) and disease progression. Toll-like (TLR) and vitamin D (VDR) receptors play important role in the immune modulation and polymorphisms that may increase CRC risk. The aim of the current study was to demonstrate the prognostic value of microbial DNA fragments in the blood of stage III CRC patients and correlate such microbial detection to TLR/VDR polymorphisms. TLR/VDR polymorphisms and presence of microbial DNA in CRC patients highlight their role in cancer development and progression.
Abstract
Gut microbial dysbiosis and microbial passage into the peripheral blood leads to colorectal cancer (CRC) and disease progression. Toll-like (TLR) and vitamin D (VDR) receptors play important role in the immune modulation and polymorphisms that may increase CRC risk and death rates. The aim of the current study was to demonstrate the prognostic value of microbial DNA fragments in the blood of stage III CRC patients and correlate such microbial detection to TLR/VDR polymorphisms. Peripheral blood was collected from 132 patients for the detection of microbial DNA fragments, and TLR/VDR gene polymorphisms. In the detection of various microbial DNA fragments, TLR and VDR polymorphisms was significantly higher compared to healthy group. Homozygous individuals of either TLR or VDR polymorphisms had significantly higher detection rates of microbial DNA fragments. Mutational and MSI status were significantly correlated with TLR9 and VDR polymorphisms. Significantly shorter disease-free survival was associated with patients with BRAF mutated tumors and ApaI polymorphisms, whereas shorter overall survival was associated with the detection of C. albicans. The detection of B. fragilis, as demonstrated by the multivariate analysis, is an independent poor prognostic factor for shorter disease-free survival. TLR/VDR genetic variants were significantly correlated with the detection of microbial fragments in the blood, and this in turn is significantly associated with tumorigenesis and disease progression.
Keywords: colorectal cancer, CRC, microbial translocation, VDR, polymorphisms, genetic variants, dysbiosis, TLR
1. Introduction
Although screening and new treatment strategies have been established for colorectal cancer (CRC), it remains a major health issue [1]. CRC is the third and second most common cancer in men and women, respectively [1,2]. Stage III CRC patients have an almost 60% 5-year overall survival rate and an almost 80% disease free survival rate [3].
CRC tumorigenesis, growth, and metastasis is a complex process, which involves molecular changes, microbial dysbiosis, impaired immunity, changes in stroma, etc. Transcriptomic profiling, microsatellite’s instability, mutation’s characteristics, somatic copy alternations number, and DNA methylation have been used as criteria for this categorization [4]. All these characteristics interact with each other and are influenced by microbial homeostasis, immune activation, or other parameters that interfere with tumorigenesis.
Microbial dysbiosis has been proven as a factor of tumorigenesis and tumor growth, especially in CRC [5,6]. Its interference with the immune system is already known [7]. Major parts of innate immunity are covered by the pattern recognition receptors (PRRs), expressed by dendritic cells and macrophages [8]. PRRs consist of Toll-like (TLRs) and NOD-like receptors. TLRs are transmembrane receptors that can detect any possible pathogens by recognizing and binding to pathogen-associated molecular patterns (PAMPs) [9]. This leads to an activation of inflammatory pathways. Their role seems to be dual in tumorigenesis [10]. TLRs 3, 5, 7, 8, and 9 enhance anti-tumor immunity through type I interferon, while TLRs 2 and 4 play a tumor-promoting role through NF-κΒ activation [11,12,13].
In recent years, the protective role of vitamin D in carcinogenesis or tumor growth is under evaluation [14,15]. Its action, especially of calcitriol, is partially regulated by the vitamin D receptors (VDRs), which are in abundance in the intestinal epithelium [16]. Palmer et al., showed that VDR expression in SW 480-ADH cells is suppressed by SNAI1 and SNAI2 transcription factors [17]. Human CRC cells have high levels of RNA encoding SNAI1 and SNAI2, leading to VDRs downregulation and interruption of vitamin D antitumor effect [17,18]. Vitamin D antitumor effect mainly interferes with proliferation, differentiation, apoptosis, angiogenesis, and immune regulation [19].
As we have previously showed, specific polymorphisms of VDRs are strongly correlated with tumorigenesis, tumor growth, and poor prognosis at stage II–IV CRC patients, the same as with TLRs polymorphisms [20,21]. TLRs polymorphisms were significant higher at CRC cancer patients than the control group and were correlated with stage IV disease and shorter overall survival [20]. Moreover, microbial translocation has been correlated to CRC tumorigenesis, metastatic disease, and shorter survival at a relevant patients group [6]. All these highlight a strong correlation between the detection of specific VDR polymorphisms, TLRs specific genetic variants, and microbial translocation, as well as the complexity of CRC. In the current study, we aimed to detect and evaluate the role of specific VDRs and TLRs polymorphisms, along with microbial translocation, in the recurrence risk of stage III CRC patients.
2. Materials and Methods
2.1. Patients and Healthy Controls Enrollment
Since August 2018 to June 2022, a total of 132 consecutive patients from the Department of Medical Oncology, University Hospital of Heraklion, aged >18 years old, with newly diagnosed and with histologically documented stage III CRC were enrolled in the study. None of the patients had history of other malignancy. Moreover, a total of 100 healthy individuals, aged >18 years old were also enrolled in the study.
2.2. Blood and Primary Tissue Samples
Peripheral blood (5 mL in EDTA) was collected from all patients and control subjects enrolled in the study, and the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany) was used for DNA extraction. Concerning the primary tissues, for microdissection of representative formalin fixed paraffin embedded (FFPE) specimens, a piezoelectric microdissector (Eppendorf, Hamburg, Germany) was used to collect cancer cells [22], following evaluation of the appropriate area by an experienced pathologist. DNA extraction was performed using both the MasterPure™ Complete DNA and RNA Purification Kit (Epicenter, Madison, WI, USA) following the manufacturer’s instructions. NanoDrop ND-1000 v3.3 (Thermo Fisher Scientific, Wilmington, DE, USA) was used for DNA quantification.
2.3. Microbial DNA Amplification
For the microbial DNA amplification, each genes’ target reagents and PCR conditions used have already been covered in detail by our team [6,23]. In brief, a set four primer pairs were used for the detection of bacterial genomic DNA encoding 16S rRNA, glutamine synthase of Bacteroides fragilis, β-galactosidase gene of Escherichia coli, and 5.8S rRNA found in Candida albicans. The samples’ DNA integrity was checked using the reference gene human glyceraldehyde phosphor-dehydrogenase (GAPDH). In order to identify bacterial DNA in blood samples, 16S rRNA was employed as a reference.
2.4. Toll-Like Receptor (TLR) and Vitamin D Receptor (VDR) Genotyping
For genotyping of TLR genetic variants, polymerase chain reaction and restriction fragment length polymorphism methodologies (PCR-RFLP) were performed. In brief, TLR2 196-to-174 Ins/Del genetic variants were determined by PCR, whereas TLR4 (Asp299Gly and Thr399Ile) and TLR9 (T1237 and T1486C) genetic variants were determined by PCR-RFLP. All materials and conditions for each gene target involved in the current study have already been described previously by others and our group [20,21,24].
For the genotyping of VDR genetic variants, as in the case of TLRs, PCR-RFLP methodology was performed. Each genes’ target reagents and PCR conditions used have already been covered in detail by our team [20,21].
In brief, for all single nucleotide polymorphisms (SNPs) of the TLR gene or VDR gene at the TaqI, ApaI, FokI, and BsmI positions, the patients were characterized as wild type, heterozygous, or homozygous in the absence of the restriction site in both alleles, the presence of the restriction site in one allele, and presence of the restriction site in both alleles, respectively.
2.5. Mutational Analysis
KRAS, NRAS, and BRAF mutational analyses were performed by Sanger sequencing, following amplification of KRAS exon 2, 3, and 4; NRAS exon 2, 3, and 4; and BRAF exon 15. Moreover, microsatellite instability (MSI) status was also evaluated. All materials and conditions for each gene target involved in the current study have already been described previously by our group [25,26,27,28].
3. Results
3.1. Patients and Healthy Donors Characteristics
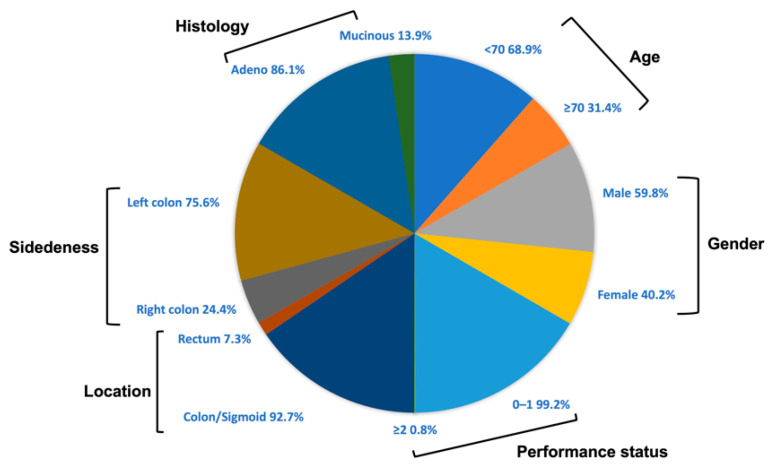
In total, 132 stage III CRC patients and 100 healthy donors have been enrolled in this single-centered study. The median age of the patients and healthy subjects enrolled was 62 years (range: 36–83 years) and 66 years (range: 35–89 years), respectively. Most of them were males (patients: 59.1%; healthy donors: 54%) and <70 years old (patients: 68.9%; healthy donors: 70%). Most of the patients had a good performance status (PS) (99.2%) and had left sided tumors (75.8%), mainly on sigmoid (50.7%). All patients were diagnosed with adenocarcinoma, and mucinous features were observed in 18 (13.9%) patients, who received CAPOX (capecitabine + oxaliplatin) as an adjuvant treatment (55.7%). The patients’ characteristics and demographics are demonstrated in Figure 1 and Table 1 and Table S1.
Figure 1.
Patients’ characteristics.
Table 1.
Patients and healthy controls characteristics.
| Demographics | Patients | Healthy Controls | ||
|---|---|---|---|---|
| Characteristics | Frequency (N = 132) | % | Frequency (N = 100) | % |
| Age median (range) | 62 (36–83) | 65 (35–89) | ||
| <70 | 91 | 68.9 | 70 | 70.0 |
| ≥70 | 41 | 31.1 | 30 | 30.0 |
| Gender | ||||
| Male | 78 | 59.1 | 54 | 54.0 |
| Female | 54 | 40.9 | 46 | 46.0 |
| Performance status | ||||
| 0–1 | 131 | 99.2 | ||
| ≥2 | 1 | 0.8 | ||
| Location | ||||
| Colon/Sigmoid | 122 | 92.4 | ||
| Rectum | 10 | 7.6 | ||
| Right/Left site | ||||
| Right colon | 32 | 24.2 | ||
| Left colon | 100 | 75.8 | ||
| Histology | ||||
| Non-Mucinous | 114 | 86.4 | ||
| Mucinous | 18 | 13.9 | ||
| Regimen | ||||
| Folfox | 59 | 44.3 | ||
| Capox | 73 | 55.7 | ||
| Treatment Duration | ||||
| 3 months | 66 | 50.0 | ||
| 6 months | 66 | 50.0 | ||
| T status | ||||
| T2 | 7 | 5.3 | ||
| T3 | 95 | 72.0 | ||
| T4 | 30 | 22.7 | ||
| N status | ||||
| N0 | 22 | 16.7 | ||
| N1 | 81 | 61.4 | ||
| N2 | 29 | 21.9 | ||
| Microsatellite Instability (MSI) | ||||
| Stable | 55 | 41.7 | ||
| High | 7 | 5.3 | ||
| Unknown | 70 | 53.0 | ||
| KRAS | ||||
| Wild type | 37 | 41.7 | ||
| Mutant | 25 | 5.3 | ||
| Unknown | 70 | 53.0 | ||
| NRAS | ||||
| Wild type | 56 | 42.4 | ||
| Mutant | 1 | 0.8 | ||
| Unknown | 75 | 56.8 | ||
| BRAF | ||||
| Wild type | 49 | 41.7 | ||
| Mutant | 5 | 5.3 | ||
| Unknown | 78 | 53.0 | ||
3.2. Detection of Microbial DNA Fragments
For both CRC patients and healthy subjects, the detection of microbial DNA fragments in the peripheral blood was evaluated. The detection of microbial fragments for 16S rRNA, E. coli, B. fragilis, and C. albicans was demonstrated in 57 (43.2%), 27 (20.5%), 42 (31.8%), and 48 (36.4%), respectively, in CRC patients, and in 16 (16%), 16 (16%), 0 (0%) and 0 (0%), respectively, in healthy controls (Table 2 and Table S1). A significant difference was demonstrated in all cases of microbial DNA fragments (p < 0.001) except for the β-galactosidase gene of E. coli (p = 0.387), between CRC patients and healthy controls (Table 2 and Table S1).
Table 2.
Association of microbial DNA fragments between CRC patients and healthy subjects.
| DNA | Gene Target | Detection | Patients | Healthy Individuals | p-Value |
|---|---|---|---|---|---|
| Microbial DNA fragments | DNA coding for 16S rRNA | Positive | 57 (43.2%) | 16 (16%) | <0.001 |
| Negative | 75 (56.8%) | 84 (84.0) | |||
| β-galactosidase gene of E. coli | Positive | 27 (20.5%) | 16 (16%) | 0.387 | |
| Negative | 105 (79.5%) | 84 (84%) | |||
| Glutamine synthase gene of B. fragilis | Positive | 42 (31.8%) | 0 (0%) | <0.001 | |
| Negative | 90 (68.2%) | 100 (100%) | |||
| DNA coding for 5.8S rRNA of C. albicans | Positive | 48 (36.4%) | 0 (0%) | <0.001 | |
| Negative | 84 (63.6%) | 100 (100%) |
3.3. TLR and VDR Genetic Variants Analysis and Clinical Outcoume
All CRC patients and healthy subjects were also evaluated for the presence of genetic variants in both VDR and TLR genes. For the case of VDR genetic variants, 20 (15.2%), 49 (37.1%), 20 (15.2%), and 17 (12.9%) patients presented the homozygous mutant genotype for TaqI, ApaI, FokI, and BsmI polymorphisms, respectively (Table 3 and Table S1). A significant difference was demonstrated for all VDR gene polymorphisms between CRC patients and healthy donors (p < 0.001) (Table 3 and Table S1). Moreover, 51 (38.6%), 52 (39.4%), 49 (37.1%), 47 (35.6%), and 75 (56.8%) patients presented the homozygous mutant genotype for TLR4—D299G, TLR4—T399I, TLR9—T1237C, TLR9—T1486C, and TLR2-196 to -174bp, respectively (Table 3 and Table S1). None of the healthy subjects presented homozygous (for all 5 TLR polymorphisms evaluated) or even heterozygous (for TLR2 and TLR4) mutant genotypes. A significant difference was also demonstrated for all TLR gene polymorphisms between CRC patients and healthy donors (p < 0.001) (Table 3 and Table S1).
Table 3.
Association of Toll-like receptors (TLRs) and vitamin D receptors (VDR) between CRC patients and healthy subjects.
| Polymorphism | Gene Target | Detection | Patients | Healthy Individuals | p-Value |
|---|---|---|---|---|---|
| VDR polymorphisms | TaqI | wild type | 55 (41.7%) | 71 (71%) | <0.001 |
| heterozygous | 57 (43.2%) | 26 (26%) | |||
| homozygous | 20 (15.2%) | 3 (3%) | |||
| ApaI | wild type | 49 (37.1%) | 52 (52%) | <0.001 | |
| heterozygous | 49 (37.1%) | 40 (40%) | |||
| homozygous | 34 (25.8%) | 8 (8%) | |||
| FokI | wild type | 41 (31.1%) | 55 (55%) | <0.001 | |
| heterozygous | 71 (53.8%) | 40 (40%) | |||
| homozygous | 20 (15.2%) | 5 (5%) | |||
| BsmI | wild type | 49 (37.1%) | 55 (55%) | <0.001 | |
| heterozygous | 66 (50%) | 43 (43%) | |||
| homozygous | 17 (12.9%) | 2 (2%) | |||
| TLR polymorphisms | TLR4—D299G | wild type | 31 (23.5%) | 100 (100%) | <0.001 |
| heterozygous | 50 (37.9%) | ||||
| homozygous | 51 (38.6%) | ||||
| TLR4—T399I | wild type | 32 (24.2%) | 100 (100%) | <0.001 | |
| heterozygous | 48 (|36.4%) | ||||
| homozygous | 52 (39.4%) | ||||
| TLR9—T1237C | wild type | 13 (9.8%) | 52 (52%) | <0.001 | |
| heterozygous | 70 (53%) | 48 (48%) | |||
| homozygous | 49 (37.1%) | ||||
| TLR9—T1486C | wild type | 13 (9.8%) | 52 (52%) | <0.001 | |
| heterozygous | 72 (54.5%) | 48 (48%) | |||
| homozygous | 47 (35.6%) | ||||
| TLR2-196 to -174bp | ins/ins | 100 (100%) | <0.001 | ||
| ins/del | 57 (43.2%) | ||||
| del/del | 75 (56.8%) |
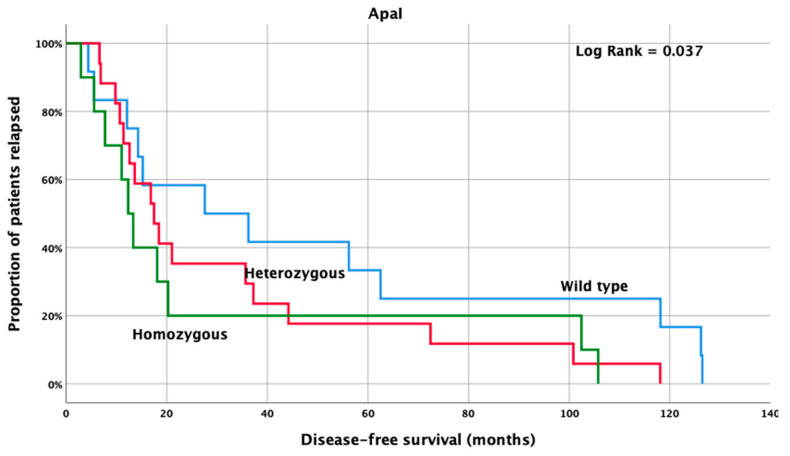
Among CRC patients, the homozygous mutant ApaI genetic variants presented with significantly lower DFS (12.3 months, 95% CI: 8.8–15.9 months) when compared to heterozygous and wild type patients (17.5 months, 95% CI: 11.0–23.9 months and 27.6 months, 95% CI: 0.0–63.2 months; p = 0.036) (Figure 2).
Figure 2.
Kaplan Meier curve for disease-free survival according to VDR-ApaI genetic variants, in stage III CRC patients.
3.4. Correlation of Microbial DNA Fragments with TLR and VDR Genetic Variants Analysis
The correlation of the presence of microbial DNA fragments, TLR, and VDR genetic variants in the peripheral blood of CRC patients were investigated (Table 4). As it was demonstrated, a significant coexistence was shown between the detection of 16S rRNA and TLR4 (D299G and T399I), TaqI and FokI polymorphisms (p = 0.009; p = 0.043; p < 0.001 and p < 0.001, respectively); E.coli was significantly associated with TaqI and ApaI polymorphisms (p < 0.001 and p = 0.003, respectively); B. fragilis was significantly associated with TLR4 (D299G) TaqI, ApaI, and FokI polymorphisms (p = 0.025; p < 0.001; p = 0.009 and p < 0.001, respectively); whereas C. albicans was significantly associated with TaqI, ApaI, FokI and BsmI polymorphisms (p < 0.001; p = 0.015; p = 0.027 and p = 0.029, respectively). Moreover, a significant association was demonstrated among the existence of almost all different TLR and VDR genetic variants (Table 4). Additionally, TLR2-196 to -174bp was significantly associated with BsmI (p = 0.004); TLR4—D299G was significantly associated with TaqI, ApaI, FokI, and BsmI (p = 0.043; p < 0.001; p = 0.015 and p < 0.001, respectively); TLR4—T399I was significantly associated with TaqI, ApaI, FokI, and BsmI (p = 0.042; p < 0.001; p = 0.036 and p < 0.001, respectively); TLR9—T1237C was significantly associated with ApaI, FokI, and BsmI (p = 0.008; p = 0.012 and p < 0.001, respectively) and TLR9—T1486C was significantly associated with ApaI, FokI, and BsmI (p = 0.018; p = 0.029 and p < 0.001, respectively).
Table 4.
Correlation between microbial DNA fragments, Toll-like receptor (TLR) and vitamin D receptor (VDR) polymorphisms parameters (values against each category represents p-values).
| Target | TLR | VDR | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Target | TLR2-196 to -174bp | TLR4—D299G | TLR4—T399I | TLR9—T1237C | TLR9—T1486C | TaqI | ApaI | FokI | BsmI |
| 16S rRNA | 0.172 | 0.009 | 0.043 | 0.549 | 0.567 | <0.001 | 0.112 | <0.001 | 0.2534 |
| Escherichia coli | 0.074 | 0.091 | 0.093 | 0.617 | 0.548 | <0.001 | 0.003 | 0.0590 | 0.553 |
| Bacteroides fragilis | 0.522 | 0.025 | 0.087 | 0.229 | 0.258 | <0.001 | 0.009 | <0.001 | 0.075 |
| Candida albicans | 0.528 | 0.798 | 0.619 | 0.896 | 0.928 | <0.001 | 0.015 | 0.027 | 0.029 |
| TLR2-196 to -174bp | <0.001 | <0.001 | <0.001 | <0.001 | 0.671 | 0.399 | 0.080 | 0.004 | |
| TLR4—D299G | <0.001 | <0.001 | <0.001 | <0.001 | 0.043 | <0.001 | 0.015 | <0.001 | |
| TLR4—T399I | <0.001 | <0.001 | <0.001 | <0.001 | 0.042 | <0.001 | 0.036 | <0.001 | |
| TLR9—T1237C | <0.001 | <0.001 | <0.001 | <0.001 | 0.364 | 0.008 | 0.012 | <0.001 | |
| TLR9—T1486C | <0.001 | <0.001 | <0.001 | <0.001 | 0.423 | 0.018 | 0.029 | <0.001 | |
| TaqI | 0.671 | 0.043 | 0.042 | 0.364 | 0.423 | 0.395 | <0.001 | <0.001 | |
| ApaI | 0.399 | <0.001 | <0.001 | 0.008 | 0.018 | 0.395 | <0.001 | <0.001 | |
| FokI | 0.080 | 0.015 | 0.036 | 0.012 | 0.027 | <0.001 | <0.001 | <0.001 | |
| BsmI | 0.003 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |
3.5. Association of Tumor Mutations and MSI Status with Microbial DNA Fragments, TLR and VDR Polymorphisms
As it is demonstrated in Table 5, the association of TLR and VDR genetic variants with RAS, RAF, and MSI status was also investigated. Of the 132 enrolled patients, KRAS, NRAS, BRAFV600E, and MSI status was available in 62 (47%), 57 (43.2%), 54 (40.9%), and 62 (47%) CRC patients, respectively (Table S1). Of those, 25 (40.3%), 1 (1.8%), 5 (9.3%), and 7 (11.3%) were KRAS, NRAS, and BRAFV600E mutants and MSI-High, respectively (Table S1). As it was observed, KRAS mutations were significantly associated with TLR9—T1237C (p = 0.014) and TLR9—T1486C (p = 0.006) polymorphisms and BRAFV600E mutations were significantly associated with TLR9—T1486C (p = 0.045) polymorphisms, whereas MSI-High status was significantly associated with TLR9—T1237C (p = 0.012), TaqI (p = 0.025), ApaI (p = 0.047), FokI (p = 0.001), and BsmI (p < 0.001).
Table 5.
Correlation of tumor mutational (KRAS/NRAS/BRAFV600E) and microsatellite instability (MSI) status with microbial DNA fragments, TLR, and VDR gene mutant alleles (values against each category represents p-values).
| Mutation/ MSI Status |
TLR9—T1237C | TLR9—T1486C | TaqI | ApaI | FokI | BsmI |
|---|---|---|---|---|---|---|
| KRAS | 0.014 | 0.006 | ||||
| BRAFV600E | 0.045 | |||||
| MSI | 0.012 | 0.025 | 0.047 | 0.001 | <0.001 |
3.6. Univariate and Multivariate Analysis for Cox Regression Analysis
According to the Cox regression univariate analysis, BRAFV600E mutations, histology (adenocarcinoma vs. mucinous), the detection of microbial DNA encoding for glutamine synthase of B. fragilis, and the detection of ApaI mutant alleles of the VDR gene are significantly associated with a shorter disease-free survival (DFS); and the detection of microbial DNA encoding for 5.8S rRNA is significantly associated with shorter overall survival (OS) (Table 6). Based on the Cox regression multivariate analysis, adjusting for above mentioned factors, B. fragilis is a significant independent factor linked to shorter OS (HR: 33.85, 95% CI: 1.8–622.4, p = 0.018) (Table 6).
Table 6.
Univariate and multivariate Cox regression analysis for disease free (DFS) and overall (OS) survival.
| Factor | Univariate | Multivariate | ||||||
|---|---|---|---|---|---|---|---|---|
| DFS | OS | DFS | OS | |||||
| Factor | HR (Range) | p-Value | HR (Range) | p-Value | HR (Range) | p-Value | HR (Range) | p-Value |
| BRAF mut vs. wt | 17.05 (2.4–123.4) | 0.005 | - | - | ||||
| B. fragilis pos vs. neg | 2.09 (1.0–4.3) | 0.047 | - | - | 33.85 (1.8–622.4) | 0.018 | - | - |
| C. albicans pos vs. neg | - | - | 3.57 (1.2–10.3) | 0.019 | ||||
| VDR—ApaI | 1.56 (1.0–2.3) | 0.031 | - | - | ||||
| Histology adeno vs. mucinus | 2.72 (1.1–6.8) | 0.031 | - | - | ||||
4. Discussion
Different pathways have been under research to understand tumorigenesis and develop new treatments against CRC. TLRs, microbiota, and VDRs are some of the areas that are under evaluation, involved in pathways that modulate immunity against cancer. In the current research, we aimed to evaluate the existence of TLRs and VDRs polymorphisms and of microbial translocation and their correlation with prognosis, in stage III CRC patients.
The passage of intestinal microorganisms into the bloodstream (also known as microbial translocation) is a phenomenon mainly met because of microbial dysbiosis [23]. This disturbance in microbial composition has been proven as one of the ways of tumorigenesis and tumor growth in CRC [6,29]. The microbiota that has escaped to the blood stream is detected mainly by its fragments or its products [6]. As our group has previously demonstrated, 16SrRNA, E. coli, B. fragilis, and 5.8S rRNA microbial fragments detected are correlated with tumorigenesis and progression and may have a prognostic role in CRC patients [6]. This study confirms the previous results, focusing on stage III CRC patients. Numerically, all DNA fragments were more frequently detected in CRC patients than in the healthy donors and this detection was statistically significant, except in the case of the detection of β-galactosidase gene of E. coli.
TLRs are a part of innate immunity, contributing mainly to the recognition of external factors that could be pathogens [10]. MyD88-dependent pathway plays a crucial role in the immune reaction and CRC related to inflammation [7,10]. Previous studies have shown that different polymorphisms have been detected in CRC patients. TLR 3/4/7/8/9 and their polymorphisms seem to have a prognostic role for CRC [20,30]. Specifically, high expression of TLR4-mediated MyD88 signaling has been correlated with poor prognosis, even in stroma and CAFs related to CRC [31,32]. Also, TLR4 along with TLR2 and TLR3 may have a prognostic role for CRC through regulation of NFκΒ pathway, leading to tumorigenesis [33]. TLR7 and TLR8 expressed by CD133+ cells have been linked to worse prognosis in CRC patients [34]. On the other hand, there are some controversial results concerning TLRs and their prognosis in various tumor types [35,36]. Specifically for CRC, high expression of TLR5 in tissue seems to be linked with better prognosis [37]. On a previous study of our group, it was demonstrated that higher frequencies of TLR2, TLR4, and TLR9 polymorphisms in CRC patients, in comparison to healthy individuals, are correlated with worst prognosis [20]. TLR2-196 to-174 del/del genotype, TLR4 Asp299Gly, TLR4 Thr399Ile, TLR9 T1237C, and TLR9 T1486C homozygous genotypes were all detected in statistically significant higher levels in the disease setting and were also correlated with worst prognosis [20]. All the above results are in accordance with the results of the current study; that is, higher rates of all TLR genetic variants detected in CRC patients compared to health individuals. Particularly for the homozygous mutant genotype for TLR4—D299G, TLR4—T399I, TLR9—T1237C, TLR9—T1486C, and TLR2-196 to-174 bp the detection was up to 56.8% in CRC patients, while none of healthy donors were detected with any of the variations. Nevertheless, no statistically significant prognostic value was demonstrated in stage III CRC patients.
The multiple roles vitamin D plays in carcinogenesis, protection, or therapy on CRC have been being researched over the last handful of decades [19]. Vitamin D fulfills its role by binding to its receptors, so VDR polymorphisms could affect the signaling pathway vitamin D activates [38]. More than 60 single nucleotide polymorphisms (SNPs) of VDR gene have been referred to in previous studies as related to carcinogenesis and prognosis at different tumor types [39]. The SNPs are found mainly in the promoter region in exons 2–9 and in the 3′-UTR (3′-untranslated regions) of the gene [40]. However, only some of them are directly related and functionally important in CRC. These include TaqI (rs731236; Thermus aquaticus I), ApaI (rs7975232; Acetobacter pasteurianus sub. pasteurianus I), BsmI (rs1544410, Bacillus stearothermophilus I), and FokI (rs2228570; Flavobacterium okeanokoites I), located in exon 9, in the intron between exons 8 and 9 and in exon 2, respectively [41,42,43,44,45]. Previous studies have highlighted the role of VDRs in immune modulation mainly through regulation of gut microbiota and microbial translocation. VDR conditional knockout (VDRΔIEC) in the epithelium of colon or low intestinal VDR protein levels may lead to microbial translocation [46]. Reduction of JAK/STAT (Janus kinases/signal transducer and activator of transcription proteins) signaling is another form of interference with gut microbiota and inflammatory responses [47]. Also, vitamin D has a promoting role for TLRs through binding to VDRs, leading to activation of innate immunity and modulation of gut microorganisms [48,49]. Different studies have tried to evaluate the role of VDR polymorphisms in CRC, with many controversial results, possibly because of the investigation of different populations [50,51,52]. Nevertheless, meta-analyses and reviews have shown a significantly higher level of detection of the homozygous mutant genotypes of TaqI and ApaI genotypes in CRC patients [53], whereas CRC tumorigenesis was correlated strongly with BsmI, FokI, and TaqI polymorphisms [54]. We have previously showed that the homozygous mutant genotypes of all TaqI, ApaI, BsmI, and FokI are significantly more frequent in CRC patients of all stages comparing to healthy donors [21]. However, the detection was in lower levels in early stages [21]. Herein, we validated the results above. Although the homozygosity of the polymorphisms were not so frequent, even in CRC patients, all evaluated polymorphisms were significantly more frequently detected in stage III CRC patients compared to the healthy population. This strongly suggests that VDR polymorphisms can contribute to CRC tumorigenesis. Regarding their correlation with TLR polymorphisms, almost all VDR and TLR polymorphisms were significantly correlated, promoting the hypothesis of their synergy. Correlating the detection of microbial fragments with VDR and TLR polymorphisms in stage III CRC patients, not all detected fragments were significantly correlated almost every polymorphism, and this is in contrast to our previous study. However, the correlation remains high, mainly for VDR polymorphisms and TLR4—D299G. All these results strengthen the hypothesis that VDR and TLR polymorphisms act together for the remodeling of gut microbiota and dysbiosis, disturbing the homeostasis and leading to immune modulation.
Finally, taking for granted the prognostic significance of tumor mutational status, KRAS and BRAF mutations were correlated with TLR9—T1237C, TLR9—T1486C polymorphisms, and TLR9—T1486C, respectively. MSI high status was correlated mainly with TLR9—T1237C and all VDR polymorphisms. Having in mind that TLR9 participates in CRC tumorigenesis through inflammation, the results can be biologically reasonable [55,56,57].
To our knowledge, our research remains the only effort to investigate the detection and possible significance for CRC tumorigenesis and progression of microbial translocation, TLR and VDR polymorphisms, and their correlation in stage III CRC patients. This research is prospective, includes a homogeneous population, is well-distributed, and took into consideration important pathological features. However, limitations do exist. The sample is relatively small and the total time of follow up needs to be extended for safer prognostic results. Despite the limitations, TLR and VDR polymorphisms, as well as microbial translocation, seem to maintain an important role to CRC tumorigenesis and progression in stage III CRC patients and could modulate immune reaction to CRC. On that basis, more research remains to be done to validate these results and drive to new, multi-targeted preventive and therapeutic options.
5. Conclusions
In conclusion, our study remains the first attempt to evaluate the detection and possible prognostic significance of TLR and VDR polymorphisms and microbial translocation in the Greek population in stage III CRC patients. All polymorphisms were significantly more frequently detected in CRC patients than in healthy donors, and the same was observed for microbial fragments with the exception of the β-galactosidase gene of E. coli. Also, these parameters seem to correlate each other, empowering the hypothesis that immune modulation against CRC is a complex axis with possible multiple preventive and therapeutical targets.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14184407/s1, Table S1: Raw data material.
Author Contributions
Conceptualization, J.S.; methodology, I.M., E.B., K.V., M.T. and M.S.; validation, I.M., M.S. and M.T.; data curation I.M., A.K., M.K., N.G., E.A., J.T., E.X., D.M., M.T. and J.S.; formal analysis, I.M.; supervision, I.M. and J.S.; writing—original draft preparation, I.M. and A.K.; writing—review and editing, I.M. and J.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study has been approved by the Ethics Committee/Institutional Review Board of the University Hospital of Heraklion (Number 7302/19-8-2009). All the procedures performed were in accordance with the ethical standards of the institutional and/or national research committee and the 1964 Helsinki declaration, and its later amendments or comparable ethical standards.
Informed Consent Statement
All patients signed a written informed consent form for their participation.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was partially funded by the Hellenic Society of Medical Oncology (HeSMO) (No: 7387/19-02-2019) and the Gastrointestinal cancer study group (GIC-SG).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Pineros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: Globocan sources and methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Yoon H.H., Shi Q., Alberts S.R., Goldberg R.M., Thibodeau S.N., Sargent D.J., Sinicrope F.A. Racial differences in braf/kras mutation rates and survival in stage iii colon cancer patients. J. Natl. Cancer Inst. 2015;107:djv186. doi: 10.1093/jnci/djv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinicrope F.A., Shi Q., Smyrk T.C., Thibodeau S.N., Dienstmann R., Guinney J., Bot B.M., Tejpar S., Delorenzi M., Goldberg R.M., et al. Molecular markers identify subtypes of stage iii colon cancer associated with patient outcomes. Gastroenterology. 2015;148:88–99. doi: 10.1053/j.gastro.2014.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alhinai E.A., Walton G.E., Commane D.M. The role of the gut microbiota in colorectal cancer causation. Int. J. Mol. Sci. 2019;20:5295. doi: 10.3390/ijms20215295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messaritakis I., Vogiatzoglou K., Tsantaki K., Ntretaki A., Sfakianaki M., Koulouridi A., Tsiaoussis J., Mavroudis D., Souglakos J. The prognostic value of the detection of microbial translocation in the blood of colorectal cancer patients. Cancers. 2020;12:1058. doi: 10.3390/cancers12041058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koulouridi A., Messaritakis I., Gouvas N., Tsiaoussis J., Souglakos J. Immunotherapy in solid tumors and gut microbiota: The correlation-a special reference to colorectal cancer. Cancers. 2020;13:43. doi: 10.3390/cancers13010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeromski J., Kaczmarek M., Boruczkowski M., Kierepa A., Kowala-Piaskowska A., Mozer-Lisewska I. Significance and role of pattern recognition receptors in malignancy. Arch. Immunol. Ther. Exp. 2019;67:133–141. doi: 10.1007/s00005-019-00540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai Z., Zhang J., Wu Q., Fang H., Shi C., Li Z., Lin C., Tang D., Wang D. Intestinal microbiota: A new force in cancer immunotherapy. Cell Commun. Signal. 2020;18:1–16. doi: 10.1186/s12964-020-00599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sameer A.S., Nissar S. Toll-like receptors (tlrs): Structure, functions, signaling, and role of their polymorphisms in colorectal cancer susceptibility. Biomed. Res. Int. 2021;2021:1157023. doi: 10.1155/2021/1157023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 12.Takeda K., Akira S. Toll-like receptors. Curr. Protoc. Immunol. 2015;109:14.12.1–14.12.10. doi: 10.1002/0471142735.im1412s109. [DOI] [PubMed] [Google Scholar]
- 13.Pradere J.P., Dapito D.H., Schwabe R.F. The yin and yang of toll-like receptors in cancer. Oncogene. 2014;33:3485–3495. doi: 10.1038/onc.2013.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deeb K.K., Trump D.L., Johnson C.S. Vitamin d signalling pathways in cancer: Potential for anticancer therapeutics. Nat. Rev. Cancer. 2007;7:684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- 15.Feldman D., Krishnan A.V., Swami S., Giovannucci E., Feldman B.J. The role of vitamin d in reducing cancer risk and progression. Nat. Rev. Cancer. 2014;14:342–357. doi: 10.1038/nrc3691. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., Zhu J., DeLuca H.F. Where is the vitamin d receptor? Arch. Biochem. Biophys. 2012;523:123–133. doi: 10.1016/j.abb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Palmer H.G., Larriba M.J., Garcia J.M., Ordonez-Moran P., Pena C., Peiro S., Puig I., Rodriguez R., de la Fuente R., Bernad A., et al. The transcription factor snail represses vitamin d receptor expression and responsiveness in human colon cancer. Nat. Med. 2004;10:917–919. doi: 10.1038/nm1095. [DOI] [PubMed] [Google Scholar]
- 18.Larriba M.J., Bonilla F., Munoz A. The transcription factors snail1 and snail2 repress vitamin d receptor during colon cancer progression. J. Steroid Biochem. Mol. Biol. 2010;121:106–109. doi: 10.1016/j.jsbmb.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Dou R., Ng K., Giovannucci E.L., Manson J.E., Qian Z.R., Ogino S. Vitamin d and colorectal cancer: Molecular, epidemiological and clinical evidence. Br. J. Nutr. 2016;115:1643–1660. doi: 10.1017/S0007114516000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messaritakis I., Stogiannitsi M., Koulouridi A., Sfakianaki M., Voutsina A., Sotiriou A., Athanasakis E., Xynos E., Mavroudis D., Tzardi M., et al. Evaluation of the detection of toll-like receptors (tlrs) in cancer development and progression in patients with colorectal cancer. PLoS ONE. 2018;13:e0197327. doi: 10.1371/journal.pone.0197327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messaritakis I., Koulouridi A., Sfakianaki M., Vogiatzoglou K., Gouvas N., Athanasakis E., Tsiaoussis J., Xynos E., Mavroudis D., Tzardi M., et al. The role of vitamin d receptor gene polymorphisms in colorectal cancer risk. Cancers. 2020;12:1379. doi: 10.3390/cancers12061379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harsch M., Bendrat K., Hofmeier G., Branscheid D., Niendorf A. A new method for histological microdissection utilizing an ultrasonically oscillating needle: Demonstrated by differential mrna expression in human lung carcinoma tissue. Am. J. Pathol. 2001;158:1985–1990. doi: 10.1016/S0002-9440(10)64669-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koulouridi A., Messaritakis I., Theodorakis E., Chondrozoumaki M., Sfakianaki M., Gouvas N., Tsiaoussis J., Mavroudis D., Tzardi M., Souglakos J. Detection of circulating tumor cells and microbial DNA fragments in stage iii colorectal cancer patients under three versus six months of adjuvant treatment. Cancers. 2021;13:3552. doi: 10.3390/cancers13143552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tahara T., Arisawa T., Wang F., Shibata T., Nakamura M., Sakata M., Hirata I., Nakano H. Toll-like receptor 2 -196 to 174del polymorphism influences the susceptibility of japanese people to gastric cancer. Cancer Sci. 2007;98:1790–1794. doi: 10.1111/j.1349-7006.2007.00590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Souglakos J., Philips J., Wang R., Marwah S., Silver M., Tzardi M., Silver J., Ogino S., Hooshmand S., Kwak E., et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br. J. Cancer. 2009;101:465–472. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sfakianaki M., Papadaki C., Tzardi M., Trypaki M., Alam S., Lagoudaki E.D., Messaritakis I., Zoras O., Mavroudis D., Georgoulias V., et al. Loss of lkb1 protein expression correlates with increased risk of recurrence and death in patients with resected, stage ii or iii colon cancer. Cancer Res. Treat. 2019;51:1518–1526. doi: 10.4143/crt.2019.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sfakianaki M., Papadaki C., Tzardi M., Trypaki M., Manolakou S., Messaritakis I., Saridaki Z., Athanasakis E., Mavroudis D., Tsiaoussis J., et al. Pkm2 expression as biomarker for resistance to oxaliplatin-based chemotherapy in colorectal cancer. Cancers. 2020;12:2058. doi: 10.3390/cancers12082058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koulouridi A., Karagianni M., Messaritakis I., Sfakianaki M., Voutsina A., Trypaki M., Bachlitzanaki M., Koustas E., Karamouzis M.V., Ntavatzikos A., et al. Prognostic value of kras mutations in colorectal cancer patients. Cancers. 2022;14:3320. doi: 10.3390/cancers14143320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kane T.D., Alexander J.W., Johannigman J.A. The detection of microbial DNA in the blood: A sensitive method for diagnosing bacteremia and/or bacterial translocation in surgical patients. Ann. Surg. 1998;227:1–9. doi: 10.1097/00000658-199801000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giordano C., Mojumdar K., Liang F., Lemaire C., Li T., Richardson J., Divangahi M., Qureshi S., Petrof B.J. Toll-like receptor 4 ablation in mdx mice reveals innate immunity as a therapeutic target in duchenne muscular dystrophy. Hum. Mol. Genet. 2015;24:2147–2162. doi: 10.1093/hmg/ddu735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunner R., Jensen-Jarolim E., Pali-Scholl I. The abc of clinical and experimental adjuvants—A brief overview. Immunol. Lett. 2010;128:29–35. doi: 10.1016/j.imlet.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eiro N., Gonzalez L., Gonzalez L.O., Fernandez-Garcia B., Andicoechea A., Barbon E., Garcia-Muniz J.L., Vizoso F.J. Toll-like receptor-4 expression by stromal fibroblasts is associated with poor prognosis in colorectal cancer. J. Immunother. 2013;36:342–349. doi: 10.1097/CJI.0b013e31829d85e6. [DOI] [PubMed] [Google Scholar]
- 33.Slattery M.L., Herrick J.S., Bondurant K.L., Wolff R.K. Toll-like receptor genes and their association with colon and rectal cancer development and prognosis. Int. J. Cancer. 2012;130:2974–2980. doi: 10.1002/ijc.26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoque R., Farooq A., Malik A., Trawick B.N., Berberich D.W., McClurg J.P., Galen K.P., Mehal W. A novel small-molecule enantiomeric analogue of traditional (−)-morphinans has specific tlr9 antagonist properties and reduces sterile inflammation-induced organ damage. J. Immunol. 2013;190:4297–4304. doi: 10.4049/jimmunol.1202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hishida A., Matsuo K., Goto Y., Naito M., Wakai K., Tajima K., Hamajima N. No associations of toll-like receptor 2 (tlr2) -196 to -174del polymorphism with the risk of helicobacter pylori seropositivity, gastric atrophy, and gastric cancer in japanese. Gastric Cancer. 2010;13:251–257. doi: 10.1007/s10120-010-0567-y. [DOI] [PubMed] [Google Scholar]
- 36.Liu S., Wang X., Shi Y., Han L., Zhao Z., Zhao C., Luo B. Toll-like receptor gene polymorphisms and susceptibility to epstein-barr virus-associated and -negative gastric carcinoma in northern china. Saudi J. Gastroenterol. 2015;21:95–103. doi: 10.4103/1319-3767.153832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beilmann-Lehtonen I., Hagstrom J., Mustonen H., Koskensalo S., Haglund C., Bockelman C. High tissue tlr5 expression predicts better outcomes in colorectal cancer patients. Oncology. 2021;99:589–600. doi: 10.1159/000516543. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C., Zhao L., Ma L., Lv C., Ding Y., Xia T., Wang J., Dou X. Vitamin d status and expression of vitamin d receptor and ll-37 in patients with spontaneous bacterial peritonitis. Dig. Dis. Sci. 2012;57:182–188. doi: 10.1007/s10620-011-1824-6. [DOI] [PubMed] [Google Scholar]
- 39.Fuhrman B.J., Freedman D.M., Bhatti P., Doody M.M., Fu Y.P., Chang S.C., Linet M.S., Sigurdson A.J. Sunlight, polymorphisms of vitamin d-related genes and risk of breast cancer. Anticancer Res. 2013;33:543–551. [PMC free article] [PubMed] [Google Scholar]
- 40.Perna L., Hoffmeister M., Schottker B., Arndt V., Haug U., Holleczek B., Burwinkel B., Ordonez-Mena J.M., Brenner H. Vitamin d receptor polymorphism and colorectal cancer-specific and all-cause mortality. Cancer Epidemiol. 2013;37:905–907. doi: 10.1016/j.canep.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Uitterlinden A.G., Fang Y., Van Meurs J.B., Pols H.A., Van Leeuwen J.P. Genetics and biology of vitamin d receptor polymorphisms. Gene. 2004;338:143–156. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Laczmanska I., Laczmanski L., Bebenek M., Karpinski P., Czemarmazowicz H., Ramsey D., Milewicz A., Sasiadek M.M. Vitamin d receptor gene polymorphisms in relation to the risk of colorectal cancer in the polish population. Tumour Biol. 2014;35:12397–12401. doi: 10.1007/s13277-014-2554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkissyan M., Wu Y., Chen Z., Mishra D.K., Sarkissyan S., Giannikopoulos I., Vadgama J.V. Vitamin d receptor foki gene polymorphisms may be associated with colorectal cancer among african american and hispanic participants. Cancer. 2014;120:1387–1393. doi: 10.1002/cncr.28565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai Y.H., Lu H., Hong D., Lin C.C., Yu Z., Chen B.C. Vitamin d receptor gene polymorphisms and colorectal cancer risk: A systematic meta-analysis. World J. Gastroenterol. 2012;18:1672–1679. doi: 10.3748/wjg.v18.i14.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jenab M., McKay J., Bueno-de-Mesquita H.B., van Duijnhoven F.J., Ferrari P., Slimani N., Jansen E.H., Pischon T., Rinaldi S., Tjonneland A., et al. Vitamin d receptor and calcium sensing receptor polymorphisms and the risk of colorectal cancer in european populations. Cancer Epidemiol. Biomark. Prev. 2009;18:2485–2491. doi: 10.1158/1055-9965.EPI-09-0319. [DOI] [PubMed] [Google Scholar]
- 46.Wu S., Zhang Y.G., Lu R., Xia Y., Zhou D., Petrof E.O., Claud E.C., Chen D., Chang E.B., Carmeliet G., et al. Intestinal epithelial vitamin d receptor deletion leads to defective autophagy in colitis. Gut. 2015;64:1082–1094. doi: 10.1136/gutjnl-2014-307436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun J. The role of vitamin d and vitamin d receptors in colon cancer. Clin. Transl. Gastroenterol. 2017;8:e103. doi: 10.1038/ctg.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arji N., Busson M., Iraqi G., Bourkadi J.E., Benjouad A., Bouayad A., Mariaselvam C., Salah S., Fortier C., Amokrane K., et al. Genetic diversity of tlr2, tlr4, and vdr loci and pulmonary tuberculosis in moroccan patients. J. Infect. Dev. Ctries. 2014;8:430–440. doi: 10.3855/jidc.3820. [DOI] [PubMed] [Google Scholar]
- 49.Liu P.T., Stenger S., Li H., Wenzel L., Tan B.H., Krutzik S.R., Ochoa M.T., Schauber J., Wu K., Meinken C., et al. Toll-like receptor triggering of a vitamin d-mediated human antimicrobial response. Science. 2006;311:1770–1773. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 50.Li C., Li Y., Gao L.B., Wang Y.Y., Zhou B., Lv M.L., Lu H.M., Zhang L. Vitamin d receptor gene polymorphisms and the risk of colorectal cancer in a chinese population. Dig. Dis. Sci. 2009;54:634–639. doi: 10.1007/s10620-008-0375-y. [DOI] [PubMed] [Google Scholar]
- 51.Yaylim-Eraltan I., Arzu Ergen H., Arikan S., Okay E., Ozturk O., Bayrak S., Isbir T. Investigation of the vdr gene polymorphisms association with susceptibility to colorectal cancer. Cell Biochem. Funct. 2007;25:731–737. doi: 10.1002/cbf.1386. [DOI] [PubMed] [Google Scholar]
- 52.Theodoratou E., Farrington S.M., Tenesa A., McNeill G., Cetnarskyj R., Barnetson R.A., Porteous M.E., Dunlop M.G., Campbell H. Modification of the inverse association between dietary vitamin d intake and colorectal cancer risk by a foki variant supports a chemoprotective action of vitamin d intake mediated through vdr binding. Int. J. Cancer. 2008;123:2170–2179. doi: 10.1002/ijc.23769. [DOI] [PubMed] [Google Scholar]
- 53.Serrano D., Gnagnarella P., Raimondi S., Gandini S. Meta-analysis on vitamin d receptor and cancer risk: Focus on the role of taqi, apai, and cdx2 polymorphisms. Eur. J. Cancer Prev. 2016;25:85–96. doi: 10.1097/CEJ.0000000000000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gandini S., Gnagnarella P., Serrano D., Pasquali E., Raimondi S. Vitamin d receptor polymorphisms and cancer. Adv. Exp. Med. Biol. 2014;810:69–105. doi: 10.1007/978-1-4939-0437-2_5. [DOI] [PubMed] [Google Scholar]
- 55.Luo Q., Zeng L., Tang C., Zhang Z., Chen Y., Zeng C. Tlr9 induces colitis-associated colorectal carcinogenesis by regulating nf-kappab expression levels. Oncol. Lett. 2020;20:110. doi: 10.3892/ol.2020.11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao C., Qiao T., Zhang B., Yuan S., Zhuang X., Luo Y. Tlr9 signaling activation at different stages in colorectal cancer and nf-kappab expression. Onco Targets Ther. 2018;11:5963–5971. doi: 10.2147/OTT.S174274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furi I., Sipos F., Germann T.M., Kalmar A., Tulassay Z., Molnar B., Muzes G. Epithelial toll-like receptor 9 signaling in colorectal inflammation and cancer: Clinico-pathogenic aspects. World J. Gastroenterol. 2013;19:4119–4126. doi: 10.3748/wjg.v19.i26.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.