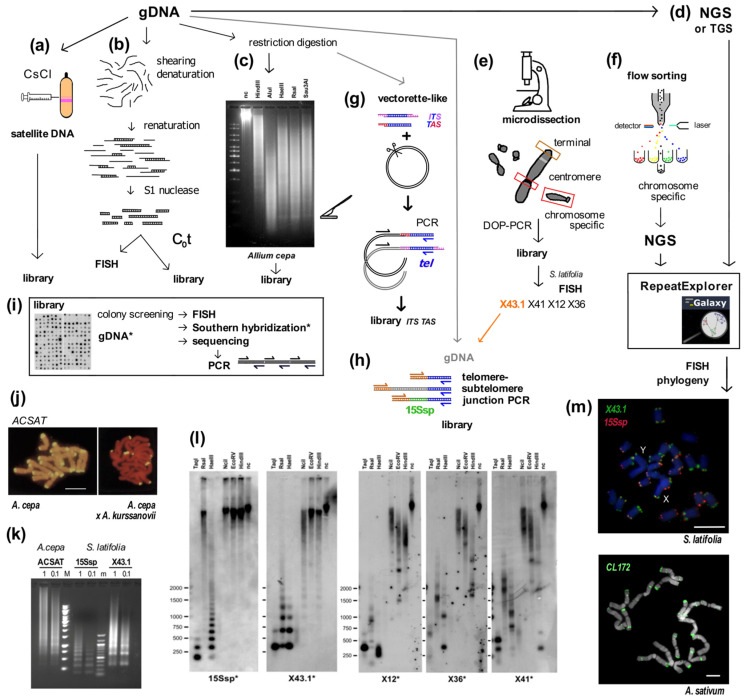
Figure 1.
Summary of experimental and in silico approaches leading to candidate satellite identification (a–h) and examples of satellite characterization (i–m). Classical experimental approaches (a–c) rely on basic physical principles. (a) Satellite DNA bands of different buoyant density are separated from the main gDNA by CsCl gradient centrifugation. (b) Single stranded DNA fragments are reassociated for a set time at high temperature to form C0t fractions. High-copy sequences bind to complementary DNA strands in short time intervals. After removal of unassociated ssDNA strands by S1 nuclease and/or hydroxylapatite chromatography, C0t fractions can be used for localization by FISH and/or for library construction. (c) Tandem repeat sequences can contain conserved sites for restriction endonucleases. After restriction digestion of gDNA, high-copy satellites form distinct bands visible on agarose gels that can be cut out and cloned. (d) gDNA samples can be used directly for Next Generation Sequencing (NGS) or Third Generation Sequencing (TGS) and data further processed in silico [39,40], e.g., using RepeatExplorer. Approaches (e,f) need state-of-the-art technologies and specific sample preparation. (e) When specific morphology parameters of chromosomes can be determined, partial or whole chromosomes can be microdissected and amplified by degenerated-oligonucleotide-primed (DOP) PCR, e.g., terminal fragment library construction from the X chromosome of dioecious plant Silene latifolia [41] (see also (l)). (f) Differences between the size of sex chromosomes and autosomes of Rumex acetosa enabled separation of chromosome-specific samples by flow cytometry [42]. (g) Vectorette PCR [43] is a genome-walking approach that enables the amplification of specific DNA fragments in situations where the sequence of only one primer is known. A vectorette-like strategy can be used to identify sequences associated with the telomere (TAS, telomere associated sequences, red) and with internal telomeric sequences (ITS, magenta). After restriction digestion of gDNA, fragments are ligated to the vector and PCR reaction with the vector-specific (black) and telomeric C-rich (blue) primer will produce a mixture of TAS and ITS sequences, e.g., identification of the subtelomeric sequence from Nicotiana tomentosiformis [44]. (h) Telomere-subtelomere junction PCR can be employed to demonstrate telomere attachment when a candidate subtelomeric sequence is known, e.g., to rDNA in Arabidopsis [45], the subtelomeric HRS60 satellite in tobacco [46] or the subtelomeric X43.1 sequence from S. latifolia [47,48]. The latter identified direct attachment of X43.1 to the telomere or via a linker formed by another satellite sequence 15Ssp (green) (see also results on (l,m)) or by various low-copy linker sequences (grey). (i) Libraries created by the approaches described above can be searched for high-copy repetitive sequences by colony hybridization using labeled (*) gDNA or C0t fraction [49,50] as a probe. Candidate clones can be used directly for localization by FISH, characterization of the genomic arrangement by Southern hybridization (examples in (l)) and/or sequenced. After sequencing, sequence-specific primers can be designed for PCR amplification of candidate sequence from gDNA (see (k)). Examples: (j) The ACSAT repeat was isolated as in (a) from Allium cepa gDNA [14] and localized by FISH (yellow) to subtelomeres on metaphase chromosomes of A. cepa (left panel). Only chromosomes of A. cepa showed ACSAT-specific signals on metaphase of an interspecific hybrid between A. cepa and A. kurssanovii (right panel) [51]. (k) Typical ladder of PCR products with periodicity corresponding to repeat unit length, demonstrating tandem organization of ACSAT, 15Ssp and X43.1 repeats when amplified with sequence-specific primers from 1 ng or 0.1 ng of gDNA of A. cepa and S. latifolia. (l) Southern hybridization demonstrated tandem organization of 15Ssp and X43.1 satellites in contrast to the dispersed pattern of X12, X36 and X41 repetitive sequences that were previously localized in subtelomeres of S. latifolia [41]. (m) FISH with LNA (locked nucleic acid) oligonucleotide probes of the X43.1 and 15Ssp satellites (upper panel, green and red, respectively) shows various localization and co-localization signals on metaphase chromosomes of S. latifolia [52]. The CL172 satellite (green, lower panel) is detected on chromosomes of A. sativum [53]. Pictures were adapted by courtesy of Prof. Ingo Schubert ((j), [51]), Dr. Terezie Mandáková ((m), [53]) and Dr. Eduard Kejnovský ((m), [52]); scale bars, 10 µm; chromosomes are counterstained with propidium iodide (j) or DAPI (m). Data in (c,k,l) are from Sýkorová (unpublished and [48]).