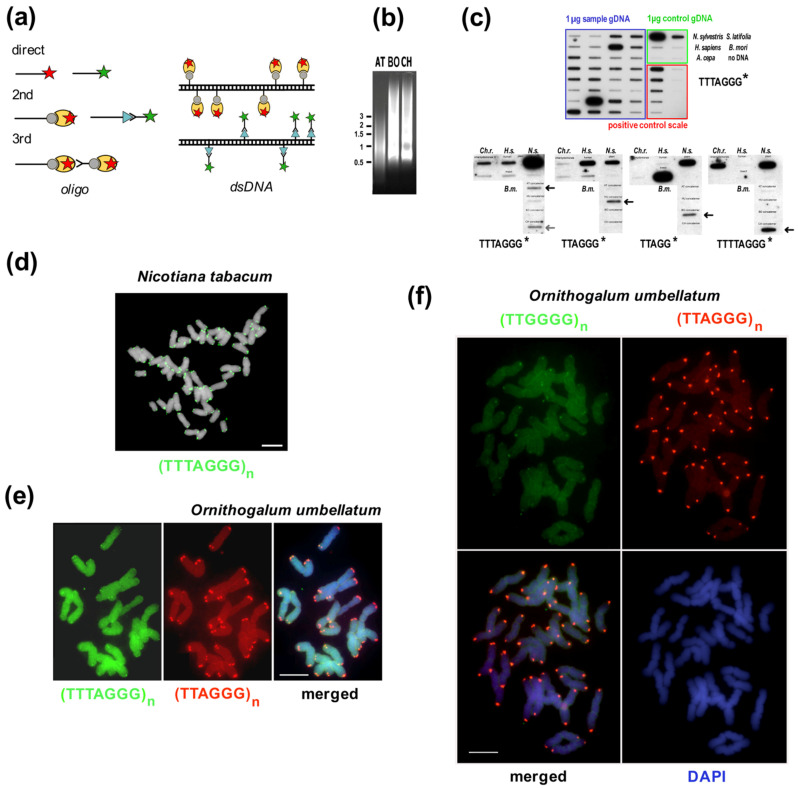
Figure 6.
Telomere identification byFISH and slot-blot hybridization. (a) Example FISH probes (top to bottom), custom synthesized oligonucleotides directly labeled with a fluorophore (asterisk). Biotinylated (circle) oligonucleotides, for secondary detection using streptavidin-bound fluorophore or tertiary detection, e.g., in a sandwich with biotinylated anti-streptavidin antibody and secondary antibody conjugated with a fluorophore. Digoxigenin-labeled (triangle) oligonucleotides are visualized by an anti-digoxigenin antibodies conjugated with a fluorophore. dsDNA probes for FISH and other hybridization techniques are often labeled in PCR reactions or by nick translation using biotin/digoxigenin modified nucleotides incorporated into products. (b) Template-free PCR produces a typical smear of products generated by self-annealing of corresponding G- and C- telomeric oligonucleotides (AT, Arabidopsis-type TTTAGGG/CCCTAAA; BO, Bombyx-type TTAGG/CCTAA; CH, Chlamydomonas-type TTTTAGGG/CCCTAAAA, [198,322]) and amplification by Taq DNA polymerase. Concatemers can be labeled radioactively for Southern hybridization or can have modified nucleotides incorporated as for FISH. (c) Slot-blot hybridization screening for telomere sequence signals using radioactively (*) labeled oligonucleotide probes. (Upper panel) gDNA samples of interest (24 species, blue frame, modified from [139]) and controls are immobilized on membrane. Control gDNA samples (green frame) and serial dilutions of control plasmids with cloned telomere sequences (red frame) serve as standards for normalization of signals between membranes when larger collections are investigated. N.B. N. sylvestris and S. latifolia are of similar genome size but differ significantly in length of telomeres. (Lower panels) Slot-blot hybridization of gDNA sets from representative species (N.s., N. sylvestris; H.s., Homo sapiens; Ch.r., Chlamydomonas reinhardtii; B.m., Bombyx mori) using four telomere probes, 100 pg of the respective concatemers (arrows) serve as a control for mutual comparison. Note crosshybridization of the oligonucleotide probe TTTAGGG to concatemers of the Chlamydomonas telomere type (grey arrow) [322]. (d) FISH of Nicotiana tabacum metaphase chromosomes using (TTTAGGG)n concatemers (green) clearly marked all termini (scale bar is 10 µm, [158]; modified). (e,f) A mixture of telomere variants occur in telomeres of Ornithogalum umbellatum. Scale bars are 5 µm. (e) The Arabidopsis-type telomeric probe (green) shows signals at some chromosome termini at incomplete metaphase of O. umbellatum and these overlap with human-type telomeric probe (red) which is dominant and represents the true telomere repeat. (f) Metaphase O. umbellatum chromosomes labeled with Tetrahymena-type concatemers ((TTGGGG)n, green) and image merged with labeled human-type concatemers (red) showing similar results to (e) ([139]; modified). All three variants of telomeric repeats occur in the Ornithogalum genome, thus FISH signals of the Arabidopsis-type (e) and Tetrahymena-type (f) variants do not result from crosshybridization and the human type was proven as the true telomere [139]. Pictures are adapted, courtesy of Dr. Terezie Mandáková (d) and Prof. Andrew Leitch (e,f), chromosomes were counterstained with DAPI.