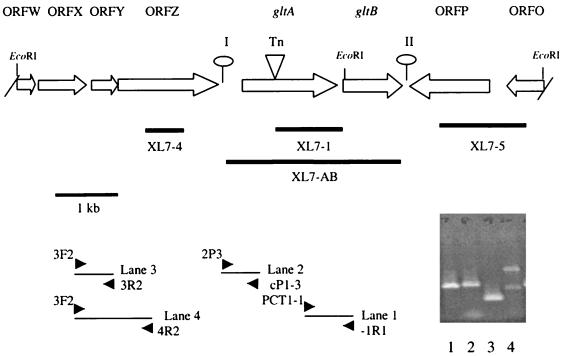
FIG. 1.
Genomic organization of the region harboring the transposon insertion in mutant XL7. Open arrows indicate ORFs and predicted direction of transcription. Slashes at the borders indicate that ORFW and ORFO are partial; lollipops represent putative stem-loop structures. The location of the Tn916ΔE insertion in gltA (Tn) is indicated by a triangle. Thick lines represent DNA fragments used as probes in Southern blots, and arrowheads at the bottom indicate primers used in RT-PCR. RT-PCR was done as described in Materials and Methods with -1R1 as the primer for cDNA synthesis. The gel shows products of PCRs with cDNA as the template and the primer pairs -1R1–PCT1-1 (lane 1), cP1-3–2P3 (lane 2), 3R2-3F2 (lane 3), and 4R2-3F2 (lane 4). Negative controls (using RNA instead of cDNA as the template and the same pairs of primers) were devoid of any product (data not shown).