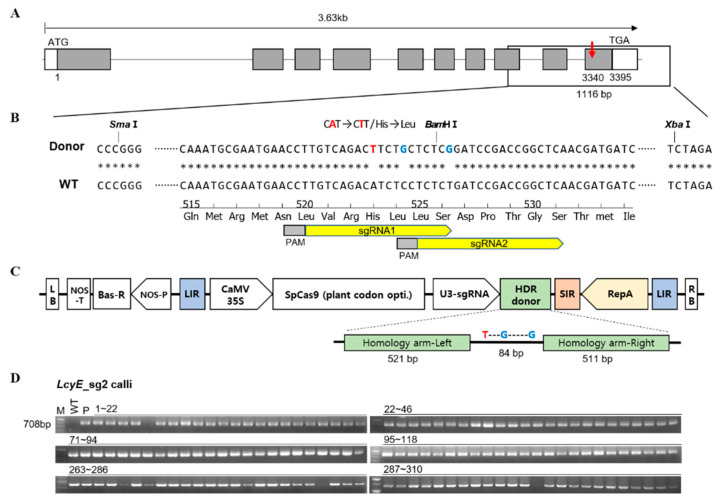
Figure 1.
(A) Schematic representation of the LcyE gene and targeting sites. (B) The nucleotides highlighted in red correspond to intended modifications for SNP, and in blue are the silent mutations to avoid re-cutting and to generate cleaved amplified polymorphic sequence (CAPS) markers. Asterisks indicate identical nucleotides. The cutting sites of the two guide RNAs used in this study are indicated by two yellow arrows with PAM. (C) Schematic representation of the CRISPR/Cas9 vector and the homologous DNA donor template construction. (D) PCR analysis of the bar gene and the NOS terminator region (primer set NOS-bar F/R) in the rice callus introduced with the pGemBos::LcyE vector. M: 1 kb DNA ladder; WT: wild type; P: pGemBos::LcyE plasmid vector.