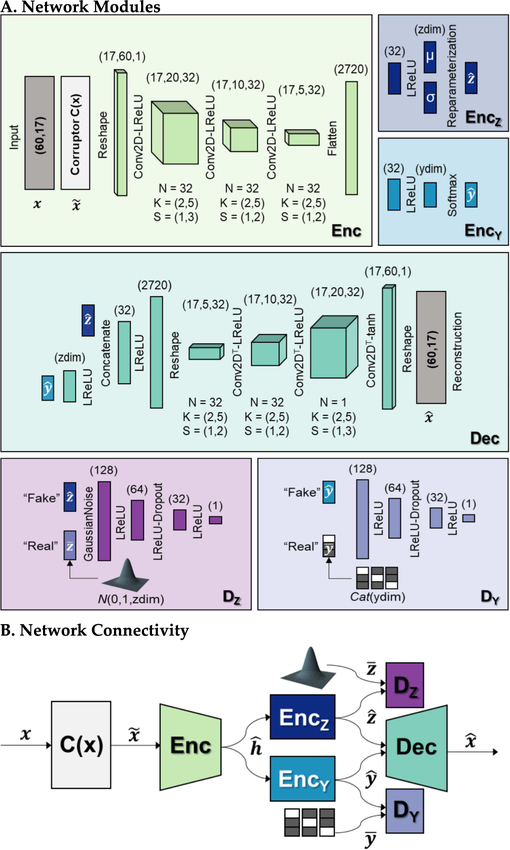
Fig. 3. Proposed deep generative model, a semi-supervised denoising adversarial autoencoder.
(A) Individual modules with output dimensions listed in parentheses above each layer. N is the number of convolution kernels, K is the kernel size, S is the stride length, LReLU represents the leaky ReLU activation, and tanh represents the hyperbolic tangent activation. The latent space dimensionality (zdim) was set to 10. “Real” samples for the latent space and label discriminators were sampled from a multivariate standard normal distribution and a categorical distribution (Cat), respectively. (B) Overall schematic of network connectivity.