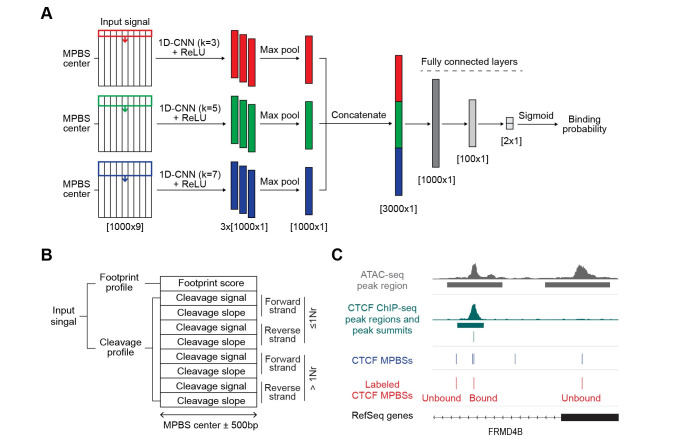
Fig 1. TAMC model and input signal.
(A) Architecture of TAMC framework: three convolutional layers with 3 learnable filters of kernel sizes k = 3, 5, and 7, and ReLU activations, followed by max pooling, concatenation, and prediction of binding probability via a two fully connected layers with sigmoid activation. (B) Default TAMC input signal structure. >1Nr, ATAC-seq read fragments larger than one nucleosome size; ≤1Nr, ATAC-seq read fragments equal or smaller than one nucleosome size. (C) Representative tracks show strategy of labeling bound and unbound CTCF MPBSs in GM12878 cell type at FRMD4B gene locus. MPBS, motif-predicted binding sites.