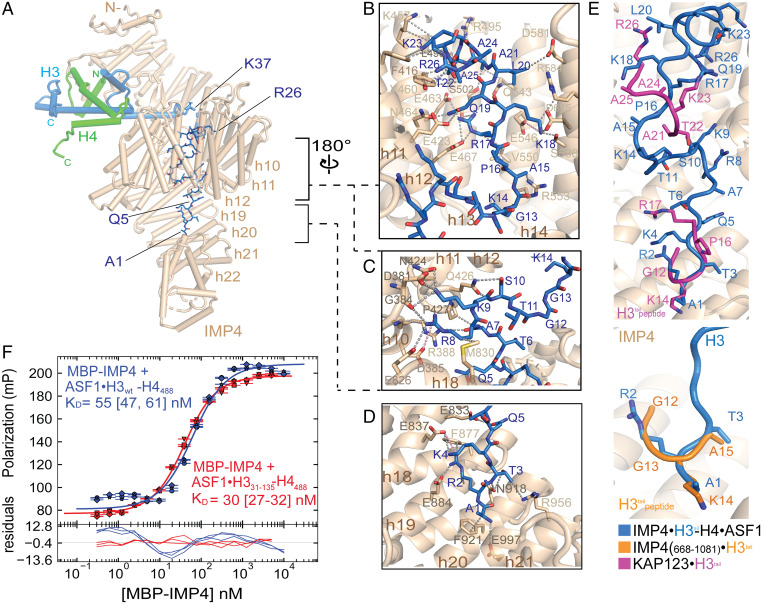
Fig. 3.
The H3tail binds the C-terminal half of IMP4. (A) A view (IMP4 is in beige, H3 is in blue, H4 is in green, and ASF1 is in yellow) showing H3tail residues 1 to 25 (blue sticks) binding to the C-terminal half of IMP4. (B–D) Details of IMP4–H3tail interactions, which mainly involve side chains (gray dashed lines) and include a few long-range electrostatic contacts (pink dashes). (B) H3tail residues 14 to 25 contact residues from HEAT repeats h11 to h15 of IMP4. (C) H3tail residues 7 to 10 bind to HEAT repeats h8 to h11 and h18. (D) H3tail residues 1 to 5 bind to repeats h18 to h22. (E) Superposition of H3tail residues of three different structures: IMP4–H3–H4–ASF1 (blue), IMP4668 to 1081–H31 to 18 (orange; PDBID: 5XBK) (22), and KAP123–H31 to 28 (magenta; PDBID: 5VE8) (12). Backbone residues of the H3tail are shown in ribbon representations, and the side chains are shown in sticks. H3tail IMP4 is beige and in cartoon representation. (F) Fluorescence polarization binding assays with MBP–IMP4 and ASF1–H3WT–H4AF488 (green line) or ASF1–H331 to 135–H4AF488 (blue line). H4 is labeled with the XFD488 fluorophore conjugated to residue 63, which is mutated to cysteine. Fitted binding curves are overlaid onto data points, with error bars representing the mean and SD of triplicate titrations. KD values are reported in the graphs with CIs of 68% in brackets.