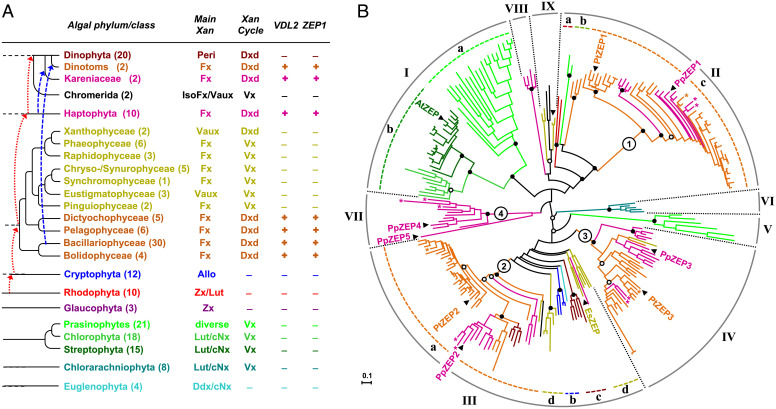
Fig. 5.
Comparative genomics of enzymes involved in fucoxanthin biosynthesis. (A) Occurrence of VDL2 and ZEP1 in publicly available sequence data from 194 species of photosynthetic eukaryotes (see Dataset S3 for species and sequence accessions). For each phylum/class, the number of examined species (in brackets), major light-harvesting xanthophyll(s) (Main Xan), and type of xanthophyll cycle (Xan cycle) are indicated. Branches indicate phylogenetic relations between taxa; dotted arrows in red and dashed arrows in blue indicate putative plastid acquisitions by endosymbioses. Allo, alloxanthin; cNx, cis-neoxanthin; Ddx, diadinoxanthin; IsoFx, isofucoxanthin; Lut, lutein; Peri, peridinin; Vaux, vaucheriaxanthin ester; Vx, violaxanthin; Zx, zeaxanthin. (B) Midpoint-rooted radial maximum-likelihood tree of the ZEP family from land plants and algae inferred from a protein alignment (324 amino acid positions) of 239 sequences from 121 species and using the same taxonomic color code as in (A) (sequences from Kareniaceae and dinotoms labeled with asterisks; see Dataset S4 for species and sequence accessions). Numbered circles indicate basal nodes of the main ZEP paralog clusters in ochrophytes and haptophytes. The paralogs from P. tricornutum (PtZEP1-3) and the haptophyte Prymnesium parvum (PpZEP1-5) and the single sequences from Arabidopsis thaliana (AtZEP) and the phaeophyte Ectocarpus siliculosus (EsZEP) are labeled. Bootstrap support (100 replicates) for major nodes indicated by black dots if higher than 90%, by white dots if higher than 75%.