Significance
The RAD51 paralog RAD51C is among the homologous recombination (HR) genes known to be tumor suppressors. As with many tumor suppressors, a large number of variants of unknown significance have been identified, impairing informed genetic counseling and treatment of cancer patients. Providing a framework for variant classification, functional analyses of RAD51C variants identify variants that confer HR deficiency and sensitize to therapeutics, including a cluster in a conserved motif. Conversely, these functional analyses support structural predictions of RAD51 paralog complexes, BDCX2 and CX3. Thus, our molecular and biochemical analyses of RAD51C variants provide insight into the functioning of the RAD51 paralog complexes.
Keywords: Homologous recombination, RAD51C, RAD51 paralog, DNA repair, Variants of unknown significance
Abstract
Mutations in homologous recombination (HR) genes, including BRCA1, BRCA2, and the RAD51 paralog RAD51C, predispose to tumorigenesis and sensitize cancers to DNA-damaging agents and poly(ADP ribose) polymerase inhibitors. However, ∼800 missense variants of unknown significance have been identified for RAD51C alone, impairing cancer risk assessment and therapeutic strategies. Here, we interrogated >50 RAD51C missense variants, finding that mutations in residues conserved with RAD51 strongly predicted HR deficiency and disrupted interactions with other RAD51 paralogs. A cluster of mutations was identified in and around the Walker A box that led to impairments in HR, interactions with three other RAD51 paralogs, binding to single-stranded DNA, and ATP hydrolysis. We generated structural models of the two RAD51 paralog complexes containing RAD51C, RAD51B-RAD51C-RAD51D-XRCC2 and RAD51C-XRCC3. Together with our functional and biochemical analyses, the structural models predict ATP binding at the interface of RAD51C interactions with other RAD51 paralogs, similar to interactions between monomers in RAD51 filaments, and explain the failure of RAD51C variants in binding multiple paralogs. Ovarian cancer patients with variants in this cluster showed exceptionally long survival, which may be relevant to the reversion potential of the variants. This comprehensive analysis provides a framework for RAD51C variant classification. Importantly, it also provides insight into the functioning of the RAD51 paralog complexes.
Faithful repair of DNA double-strand breaks (DSBs) is essential for preserving genome stability. Homologous recombination (HR) is a high-fidelity DSB repair pathway used to repair both endogenous and exogenous sources of DNA damage (1). Mutations in HR genes are associated with many cancers but are especially prevalent in breast, ovarian, and prostate cancers (2–4). For example, deleterious HR gene mutations, both germline and somatic, occur in a third to a half of ovarian carcinomas (2). Truncation mutations are consistently considered to be deleterious, but a large number of missense mutations have been identified through clinical or tumor sequencing and most are variants of unknown significance (VUSs). HR deficiency (termed HRD) sensitizes cancers to chemotherapies, e.g., crosslinking agents and poly(ADP ribose) polymerase (PARP) inhibitors, and thus impacts therapeutic strategies (5, 6).
While BRCA1 and BRCA2 are the two most frequently mutated HR genes in cancers, several other HR genes, including RAD51C, RAD51D, and PALB2, are also tumor suppressors (1). When these are mutated, the resulting HR-deficient cancers can be targeted by similar therapies as BRCA1 and BRCA2 mutant neoplasms (7, 8). Thus, screening panels and targeted tumor sequencing typically include these and other HR genes (e.g., BROCA Cancer Risk Panel, cbioportal) (2, 9–11). RAD51C mutations, including frameshifts and missense mutations, as well as promoter methylation, have been identified in 3% of ovarian cancers (2). Deleterious RAD51C mutations are constitutionally heterozygous within the patient but exhibit loss of heterozygosity (LOH) within the cancer (12). Ovarian cancer risk for RAD51C mutation carriers is on par with that of BRCA2 mutation carriers (odds ratio RAD51C of 8.3 versus BRCA2 of 9.94 [95% CI, 5.43 to 12.48 and 8.09 to 12.25, respectively] (13)). These cancers are also typically p53 deficient, which likely enables the neoplastic cells to tolerate RAD51C loss (7, 12).
The incorporation of RAD51C into clinical mutation screening panels and targeted tumor sequencing has uncovered a large number of missense mutations that are considered VUSs. This poses an impediment to assessing cancer risk for individuals harboring germ-line mutations. Similarly, RAD51C VUSs reported in cancers may affect therapy response when the wild-type (WT) copy of RAD51C has been lost. Systematic classification of RAD51C variants, therefore, would be instrumental in these cases for determining cancer risk and for predicting the response to therapies that target HR-deficient cancers.
A central step in HR is RAD51 filament formation on single-stranded DNA (ssDNA), which is derived from the processing of a primary DNA lesion, such as a DSB. RAD51-ssDNA nucleoprotein filaments perform the homology search and strand invasion steps that underpin the HR process (14). RAD51C is one of five canonical RAD51 paralogs in mammalian cells that regulate RAD51 activity (15, 16). In budding yeast and worms, Rad51 paralogs act as chaperones to accelerate the assembly of Rad51-ssDNA nucleoprotein filaments (17–20), and it is presumed that the mammalian homologs act similarly, although the components of the complexes differ. Like RAD51, the RAD51 paralogs contain conserved Walker A and B motifs for ATP binding and/or hydrolysis (16), possibly to regulate protein-protein interactions as for RAD51.
In this study, we performed an extensive analysis of 56 RAD51C variants derived from various databases (ClinVar, cbioportal, and gnomAD), most of which are predicted to be deleterious by in silico prediction algorithms. However, we observed a wide range of HR levels in the variants relative to WT RAD51C, classifying slightly less than half of the variants as HR deficient, despite predictions of pathogenicity. Thus, prediction tools were limited in their ability to assess HR function. However, even though RAD51C identity with RAD51 is limited, we found that RAD51C variants with severe HR defects are usually mutated in residues that are conserved with RAD51; conversely, HR-proficient variants typically harbor mutations at residues that are not conserved with RAD51. Defective interactions with other RAD51 paralogs were also observed and correlated well with HRD. Finally, a mutation cluster was observed around the Walker A motif; biochemical characterization of variants in or around the motif demonstrated deficiencies in ATP hydrolysis and ssDNA binding. Structural models of RAD51C in complexes with other RAD51 paralogs are presented that are supported by our functional analysis and predict ATP binding between RAD51 paralogs. Ovarian cancer patients with mutations in this cluster had exceptional long-term survival. Our comprehensive analysis of RAD51C missense variants, including structural modeling of RAD51 paralog complexes, provides insight into RAD51C variant classification that should be valuable in assessing other RAD51 paralog variants.
Results
RAD51C VUS Selection for Analysis.
Through extensive sequencing efforts, numerous RAD51C missense variants have been identified during clinical testing and cancer and population sequencing but with relatively little functional investigation as to their significance. The ClinVar database currently reports ∼600 missense variants, mostly from clinical testing, covering 86% of the 376 RAD51C amino acids, the vast majority of which are of uncertain significance (SI Appendix, Table S1). Regarding RAD51C cancer variants, ∼300 different missense variants have been reported in ∼400 cancers derived from various tissue types, many of which overlap with those identified through clinical sequencing (SI Appendix, Table S2, see below). GnomAD sequencing of various populations has identified ∼200 missense variants in ∼150 residues, most of which are rare (1 to 5 individuals; population frequency of ∼10−5 to ∼10−6), although a few are observed at a much higher frequency (∼10−3) (SI Appendix, Table S3). From these 3 sources, ∼800 unique RAD51C variants have been reported. Only ∼1% of these variants have been annotated as pathogenic or benign (SI Appendix, Table S1).
We selected 56 RAD51C variants for functional analysis, of which 45 were not previously characterized and so are considered VUSs (22 cancer variants, 5 population variants, and 18 annotated as both), together with 11 that were previously functionally characterized (4 cancer variants, 7 annotated as both) (Fig. 1A and Tables 1 and 2). The 51 RAD51C variants identified in cancers were both germline (28 variants) and somatic (23 variants) in origin (Table 1). The 45 VUSs were chosen for analysis based on several criteria, as follows: sequence conservation of the cognate residue and position within a conserved domain (SI Appendix, Fig. S1A), predictions of the variant being damaging for protein function, and/or presence in breast/ovarian cancers, given the known association of mutations in this gene with these cancers. We utilized three prediction tools, namely, PolyPhen-2, SIFT, and PROVEAN (22–24), to gauge the functional significance of the variants, giving each tool a predictive score of 0 (benign) or 1 (damaging). Nearly two-thirds of the variants (34/56, 61%) are predicted to be damaging using all three algorithms (SI Appendix, Table S4).
Fig. 1.
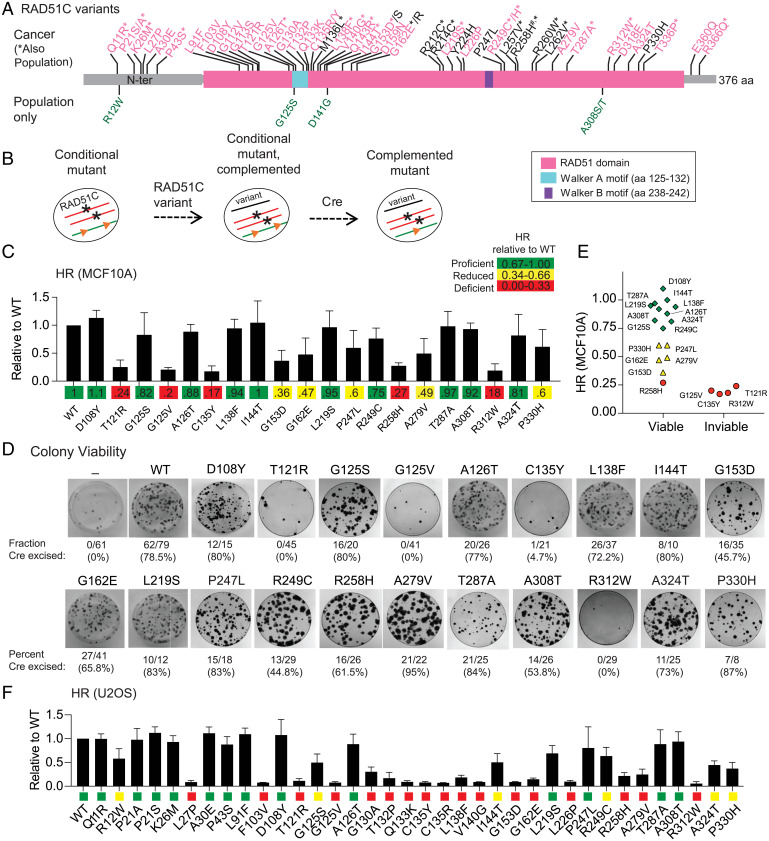
HR-deficient RAD51C variants identified using MCF10A and U2OS cell mutants. (A) RAD51C variants analyzed in this study. Cancer variants are indicated along the top of the schematic. Breast or ovarian cancer (BC/OC) variants are in pink; variants found in other cancer types are in black (see Table 1). Cancer variants that have also been identified as population variants are indicated with an asterisk; four population variants not identified in cancers are indicated under the schematic in green (see Table 2). R258H, annotated with a #, was initially identified as a biallelic mutation in Fanconi-anemia-like syndrome patients. Identified motifs in RAD51C include the Walker A and B motifs that are important for nucleotide binding and hydrolysis. aa, amino acid; N-ter, N terminus. (B) Schematic for testing RAD51C variants in MCF10A cells. Both endogenous alleles of RAD51C are disrupted (asterisks), but cells are viable due to expression from a conditional, ectopically integrated copy of RAD51C flanked by LoxP sites (orange triangles). RAD51C variants are introduced by viral transduction, and their ability to complement is determined after Cre-mediated excision of the conditional RAD51C copy. (C) HR levels in MCF10A cells expressing the indicated variants. A direct repeat HR reporter (DR-GFP) is present in the genome of the MCF10A conditional cell line. Upon introduction of a DSB in the reporter, repair by HR produces a functional GFP gene that is quantified by flow cytometry. HR levels for the variants are expressed relative to the HR level achieved with WT RAD51C. (D) MCF10A colony formation after Cre expression in cells containing the indicated RAD51C variants. The number of colonies that had excised the conditional copy of RAD51C compared to the total that were analyzed is indicated together with the percent excision. After 10 d of growth, three variants (T121R, G125V, and R312W) were unable to give rise to viable colonies, and for a fourth, C135Y, a single clone that had excised the conditional RAD51C copy, did not survive past two passages. (E) Comparison of HR and viability in MCF10A cells. RAD51C variants called as HR proficient or reduced support viability of MCF10A cells, while those called as HR deficient do not, except for R258H, which is on the higher end of HRD. (F) HR levels in RAD51C knockout U2OS cells expressing the indicated RAD51C variants. A distinct HR reporter (SCR-GFP), in which HR is also induced by a DSB, is present in the genome of the U2OS cells. HR was quantified relative to WT RAD51C and categorized as proficient (green), reduced (yellow), and deficient (red), as in C.
Table 1.
RAD51C cancer variants analyzed in this study
| Variant | Cancer type | Allele origin |
|---|---|---|
| Q11R* | Epithelial ovarian cancer | Germline# |
| P21A* | Breast carcinoma | Somatic# |
| P21S | Breast carcinoma | Somatic# |
| K26M* | Epithelial ovarian and breast cancer | Germline# |
| L27P | Epithelial ovarian cancer | Germline# |
| A30E | Breast cancer | Germline |
| P43S | Epithelial ovarian cancer | Germline# |
| L91F | Epithelial ovarian cancer | Germline# |
| F103V | Bilateral breast cancer | Germline# |
| D108Y | Colorectal adenocarcinoma | Somatic |
| G112V | Epithelial ovarian cancer | Germline# |
| G113S | Breast invasive ductal carcinoma | Somatic |
| T121R | Ovarian cancer | Somatic |
| G125V | Breast cancer (pathogenic) | Germline# |
| A126T* | Breast and ovarian cancer, melanoma (benign) | Germline# |
| G130A | Breast cancer | Somatic# |
| T132P | Ovarian cancer (likely pathogenic) | Germline# |
| Q133K | Epithelial ovarian cancer | Germline# |
| C135R | Breast cancer | Germline |
| C135Y | Breast and ovarian cancer | Germline |
| M136L* | Lung adenocarcinoma | Somatic# |
| L138F* | Breast and ovarian cancer (pathogenic) | Germline# |
| V140G* | Epithelial ovarian cancer | Germline |
| Q143R* | Breast cancer (uncertain) | Germline# |
| I144T* | Epithelial ovarian cancer | Germline# |
| G153D* | Breast cancer | Germline# |
| G153S | Liver cancer | Somatic |
| D159N | Breast cancer (conflicting) | Germline# |
| G162E* | Breast and ovarian cancer | Germline# |
| G162R | Malignant melanoma | Somatic |
| R212C* | Lung and uterine endometrioid carcinoma | Somatic# |
| R214C* | Colon adenocarcinoma | Somatic# |
| L219S* | Breast and ovarian cancer (conflicting) | Germline# |
| Y224H | Bladder urothelial carcinoma | Somatic |
| L226P | Epithelial ovarian cancer | Germline# |
| P247L | Malignant melanoma | Somatic# |
| R249C* | Breast cancer, cutaneous melanoma, duodenal adenocarcinoma | Somatic# |
| R249H* | Epithelial ovarian cancer | Germline# |
| L257V* | Liver cancer | Somatic# |
| R258H* | Colon and prostate carcinoma, GBM (pathogenic) | Somatic# |
| R260W* | Prostate adenocarcinoma | Somatic# |
| L262V* | Epithelial ovarian cancer | Germline# |
| A279V | Endometrioid carcinoma | Somatic |
| T287A* | Breast and ovarian cancer (benign) | Germline# |
| R312W* | Ovarian and large intestine carcinoma (conflicting) | Germline, somatic# |
| D318E | Breast carcinoma | Somatic |
| A324T | Colorectal adenocarcinoma | Somatic# |
| P330H | Urinary tract carcinoma | Somatic |
| T336P* | Epithelial ovarian cancer | Germline# |
| E360Q | Invasive breast carcinoma | Somatic |
| R366Q* | Breast and ovarian cancer (uncertain) | Germline# |
Sources of the cancer variants are indicated in SI Appendix, Table S1. An asterisk indicates the variant was also identified in populations (Table 2). R258H was also identified as a biallelic germ line mutation in Fanconi anemia–like syndrome. Variants also found in ClinVar, which are all germline, are indicated with a “#". The clinical significance notation from ClinVar is indicated in parentheses for the 10 variants with previous functional characterization. The rest of the variants that are in ClinVar are listed as uncertain significance, except for C135Y, which is listed as likely pathogenic, and L219S and R312W, which are listed as conflicting interpretations of pathogenicity.
Table 2.
RAD51C population variants
| Variant | Population | Allele count/total |
|---|---|---|
| Q11R | European | 1/251,450 |
| R12W | African/African American, European | 2/251,456 |
| P21A | South Asian | 1/251,466 |
| K26M | European | 2/282,878 |
| P43S | Latino/Admixed American | 1/251,412 |
| G125S | European | 1/250,004 |
| A126T | Mostly European | 981/281,388 |
| M136L | East Asian | 3/251,476 |
| L138F | European | 1/251,476 |
| V140G | European | 1/251,480 |
| D141G* | European American | 1/13,006 |
| Q143R | European | 9/251,480 |
| I144T | European | 16/282,878 |
| G153D | Latino/Admixed American, European | 2/251,462 |
| G162E | European | 1/251,482 |
| R212C | Mostly South Asian, European | 9/282,638 |
| R214C | African/African American, East Asian | 22/282,622 |
| L219S | Latino/Admixed American, European | 2/251,286 |
| R249C | European | 1/251,440 |
| R249H | Mostly African/African American | 5/251,434 |
| L257V | Mostly Latino/Admixed American | 9/251,444 |
| R258H | European | 4/251,428 |
| R260W | African/African American, East Asian | 2/251,438 |
| L262V | Mostly European | 20/282,828 |
| T287A | Mostly European | 1594/282,558 |
| A308S | African/African American | 17/282,782 |
| A308T | African/African American | 1/251,384 |
| R312W | East Asian and European | 2/251,360 |
| T336P | African/African American, European | 3/282,810 |
| R366Q | Mixed | 3/249,618 |
All RAD51C population variants were sourced from gnomAD (https://gnomad.broadinstitute.org/) except D141G (*), which is from dbSNP (https://www.ncbi.nlm.nih.gov/snp/). Note that homozygotes for A126T and T287A have been identified in 3 and 7 individuals, respectively. Allele counts were last updated December 2021.
A Subset of RAD51C VUSs Confer Defects in Sustaining Cell Viability and HR.
RAD51C is essential for the survival of the nontransformed human mammary epithelial cell line MCF10A, presumably due its role in HR (25). To investigate the impact of the variants on HR, we utilized our recently developed MCF10A RAD51C conditional cell line (conditional mutant, Fig. 1B) in which the two endogenous copies of RAD51C are disrupted (25). These cells are viable due to the expression of an additional copy of RAD51C at the AAVS1 locus that is flanked by LoxP sites, but they are inviable upon Cre-mediated recombination unless the cells are complemented by a functional RAD51C gene. Complementation by RAD51C variants is achieved by lentiviral transduction prior to Cre expression. Upon Cre expression and excision of WT RAD51C at the AAVS1 locus, the ability of each variant to support cell survival is determined (Fig. 1B). These cells also contain a stably integrated direct-repeat reporter to measure HR through the introduction of a DSB in the reporter and the appearance of GFP-positive cells following HR.
We selected 20 of the 56 RAD51C variants for HR and viability analysis in MCF10A cells. Cells were first evaluated for HR soon after Cre expression, prior to viability issues that could arise. Variants were classified relative to the WT as HR proficient (67 to 100%), HR reduced (34 to 66%), or HR deficient (0 to 33%). Half of the variants were found to be either HR deficient or HR reduced, whereas the remaining half were HR proficient (Fig. 1C). We then determined which variants would permit survival in MCF10A cells. As expected, viable colonies that had excised the conditional RAD51C allele were obtained on plates from cells expressing WT RAD51C but not in the absence of RAD51C complementation (Fig. 1D). Of the 20 RAD51C variants tested, 16 were able to support viability while the remaining 4 were not. HR proficiency correlated with cell viability, as cells expressing 4 of the 5 variants classified as HR deficient were inviable (Fig. 1E). The one variant categorized as HR deficient and yet able to confer viability, namely, R258H, supported the highest level of HR within the HR-deficient group and so it may reflect the threshold of HR activity required for viability. Consistent with its ability to sustain cell survival, R258H has been identified as a homozygous mutation in children with a Fanconi anemia–like syndrome (26).
Unlike nontransformed MCF10A cells, RAD51C is not essential in the osteosarcoma cell line U2OS (25), allowing rapid testing of additional RAD51C variants for HR activity. Using an integrated GFP-based reporter, we tested 36 RAD51C variants in U2OS cells for HR proficiency, including those we had examined in MCF10A cells. We again observed a wide range of HR levels relative to WT RAD51C, categorizing 14 variants as HR proficient (39%), 5 as HR reduced (14%), and 17 as HR deficient (47%) (Fig. 1F). Western blotting confirmed the expression of all of the variants associated with defective HR (SI Appendix, Fig. S2A). Overall, the impact of RAD51C variants on HR correlated well between MCF10A and U2OS cells (r = 0.82; SI Appendix, Fig. S2B).
Interactions with Other RAD51 Paralogs Are Frequently Disrupted in RAD51C Variants.
The five canonical RAD51 paralogs interact with each other to form two complexes, namely, RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) and RAD51C-XRCC3 (CX3) (15, 16) (Fig. 2A). RAD51C is the only common member of the two complexes and directly interacts with RAD51B, RAD51D, and XRCC3. We first tested whether the RAD51C variants were compromised for these interactions using the yeast two-hybrid (Y2H) approach for RAD51B and XRCC3 and the yeast three-hybrid (Y3H) approach for RAD51D because its interaction with RAD51C is more robust in the presence of its binding partner RAD51B (27, 28).
Fig. 2.
HR-deficient RAD51C variants typically have impaired interactions with other RAD51 paralogs. (A) RAD51 paralog complexes. The BCDX2 complex contains RAD51B, RAD51C, RAD51D, and XRCC2 and the CX3 complex contains RAD51C and XRCC3. RAD51C is the sole member present in subcomplexes. (B) Representative Y2H and Y3H interactions of RAD51C variants with other RAD51 paralogs, as monitored by growth. A Y2H approach was used to test RAD51C interactions with either RAD51B or XRCC3, and a Y3H approach was used for RAD51D because the RAD51C interaction is more robust in the presence of its binding partner RAD51B. RAD51C variants were tested as here as activating domain (AD) fusions as well as reciprocal DNA binding domain (BD) fusions (SI Appendix, Fig. S3A). (C) Heat map indicates averaged protein-protein interactions as determined in the Y2H/Y3H assays. The RAD51C variant interaction with the indicated RAD51 paralog is relative to the WT. Growth was quantified using ImageJ from two or more independent experiments. The last column shows HR quantification from U2OS cells for comparison. Variants R212C and R214C were tested later than the other variants and found to be HR proficient. (D–F) HR levels for the RAD51C variants correlate well with Y2H/Y3H interactions for each of RAD51D (D), RAD51B (E), and XRCC3 (F), with the RAD51D interaction having the highest correlation the best predictor (RAD51D and RAD51B, P < 0.0001; XRCC3, P < 0.001). Linear regression lines are shown in black with 90% confidence intervals shaded in green.
In these Y2H and Y3H systems, 56 RAD51C variants were tested as an activating domain fusion (Fig. 2B) and most also as a DNA binding domain fusion (SI Appendix, Fig. S3 A–F). Just as we did in the HR analysis, Y2H/Y3H results were classified relative to WT RAD51C as proficient, reduced, or deficient (Fig. 2C). More than half of the RAD51C variants (32/56) were defective (reduced/deficient) in an interaction with one or more of the RAD51C binding partners. We found 16 variants to be specifically compromised for one or two interactions, most often involving RAD51D (15/16). In the other 16 variants, all 3 interactions (RAD51D, RAD51B, XRCC3) were scored as defective, with 14 of the amino acid changes clustered between residues F103 and G153. Thirteen of the 16 variants that scored as deficient in all 3 interactions were tested by Western blotting and verified to be expressed although in some cases at a lower level than WT RAD51C (SI Appendix, Fig. S3G).
Overall, defects in the RAD51C variant interactions with any of the three other paralogs correlated well with HRD, with disruption of the RAD51D interaction being the best correlated (r = 0.85, Fig. 2 D–F; r = 0.90; SI Appendix, Fig. S3 G–I). An exception was L27P, in which all three interactions appeared normal but HR was highly deficient (Fig. 2C). The variant R258H, originally identified in Fanconi anemia patients, was also interesting in that the interaction with RAD51D, but not with RAD51B or XRCC3, was somewhat reduced but HR was quite deficient (see below).
RAD51C Residue Conservation with RAD51 Is Predictive for Variant Function.
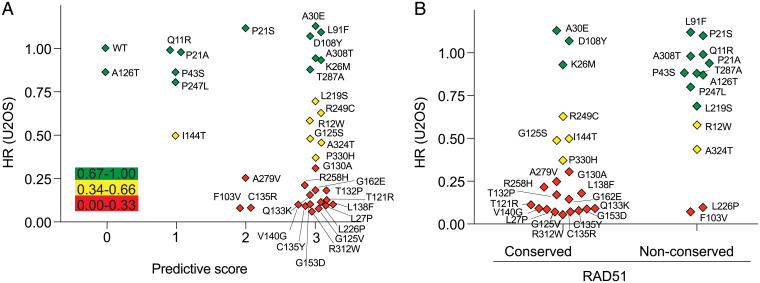
We compared HR results obtained for the RAD51C variants with the scores from three in silico prediction algorithms (PolyPhen-2, SIFT, and PROVEAN). Of the 26 RAD51C variants considered deleterious by all 3 prediction tools, 14 were scored as HR deficient (54%); 6 as HR reduced (23%), but with apparently sufficient activity to support MCF10A viability (5 of 6 tested); and 6 as HR proficient (23%) (Fig. 3A). Thus, these tools were limited in their capacity to predict HR function. As an example, T287A is predicted to be deleterious by all three prediction tools, but it is fully proficient for HR in both MCF10A and U2OS cells and supports MCF10A survival (Fig. 1 C–F). Further confirming its functionality, individuals constitutionally homozygous for T287A have been identified in population analyses (Table 2), and the protein was active in other cellular assays (12). An additional five variants were predicted to be deleterious by one or two tools, and yet they were fully HR proficient (e.g., Q11R or P21S, respectively). Conversely, three variants were scored as benign by one of the prediction tools, and yet they were HR deficient (e.g., A279V).
Fig. 3.
RAD51C residue conservation with RAD51 is predictive for variant HR function. (A) Comparison of HR activity of the RAD51C variants in U2OS cells with the combined scores from the PolyPhen-2, SIFT, and PROVEAN prediction tools. A predictive score of 0 (benign) or 1 (damaging) is given for each tool, according to the cut off score for each, and then the three scores are added. Thus, a combined score of 0 indicates that all three tools predict that the variant is benign, while a combined score of 3 indicates that all three tools predict that the variant is deleterious. The variants are colored according to whether HR is proficient (green), reduced (yellow), or deficient (red). (B) Comparison of HR activity of the RAD51C variants in U2OS cells with residue conservation with RAD51. Conserved, RAD51C amino acid is identical or strongly similar to the cognate residue in RAD51; nonconserved, RAD51C amino acid is weakly similar or not similar at all to the cognate residue in RAD51.
In addition to prediction tools for protein function, a statistical algorithm has been developed to identify recurrently mutated residues in tumor samples, thus defining cancer hot spots (www.cancerhotspots.org) (21, 29). RAD51C P21 has been annotated as a cancer hot spot. It has been reported mutated to P21A, P21S, or P21T in 18 cancers and, additionally, P21L in ClinVar. We found that P21A and P21S are both HR proficient (Fig. 1F), suggesting that these variants are unlikely relevant to the disease. It is notable that the P21 codon is in a highly GC-rich sequence within a small inverted repeat, such that the sequence context itself may be prone to mutagenesis (SI Appendix, Fig. S2C).
As a RAD51 paralog, RAD51C shares 28% sequence identity with RAD51. We asked whether variants in residues that are identical to RAD51 are more likely to be HR defective. Of the 33 RAD51C residues that were altered in the 36 variants tested for HR (30 with a single variant, 3 with 2 variants), 22 were at residues that are either identical or highly conserved with RAD51, while the other 14 were not conserved or poorly conserved (SI Appendix, Fig. S4A). In the conserved residues, 68% of variants were HR deficient, compared with 14% of variants in the nonconserved residues (P = 0.0022, Fisher’s exact test; Fig. 3B), indicating that conservation with RAD51 can be predictive for function.
Because there was a bias in choosing RAD51C variants for HR analysis, i.e., based on their performance in Y2H and Y3H assays or their position within a conserved domain within the protein, we also compared the total score from the 3 predictive algorithms with the RAD51D interaction, which had been tested for all 56 RAD51C variants and which shows good correlation to HR proficiency. Of the 34 RAD51C variants expected to be damaging by all 3 prediction algorithms, only 17 (50%) were deficient in the RAD51D interaction, whereas 11 (33%) were proficient for the interaction and 6 (17%) were partially impaired for the interaction (SI Appendix, Fig. S4B). Thus, the tools were again limited in their predictive ability. We also considered effects on the RAD51D interaction in the context of whether the residue that was mutated is conserved with RAD51. In the conserved residues, 17 of 33 variants (52%) were deficient in the RAD51D interaction, compared with 5 of 23 variants (22%) in nonconserved residues (P = 0.0298, Fisher’s exact test) (SI Appendix, Fig. S4C). Thus, variants in residues that are not conserved with RAD51 were likely to maintain the RAD51D interaction and HR proficiency.
Identification of a Mutation Cluster in the Walker A Region.
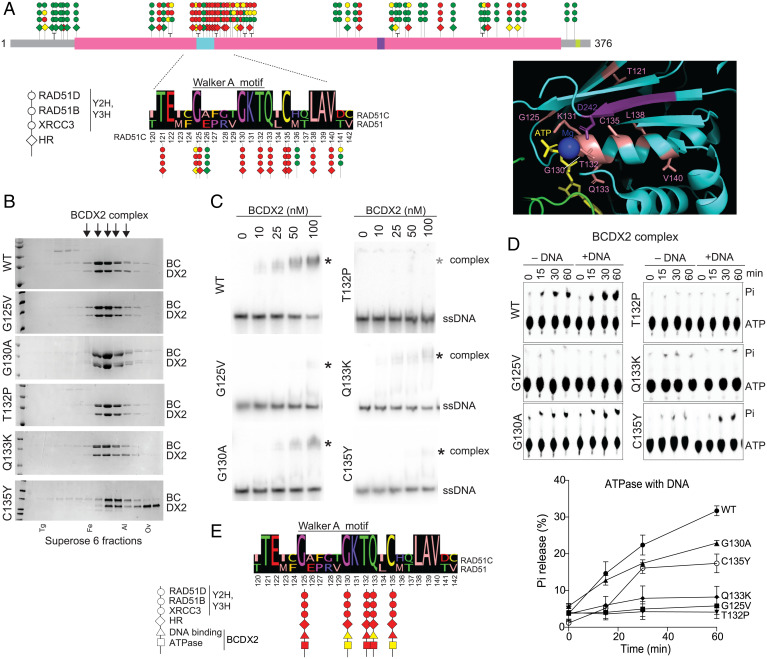
We compiled our HR and protein interaction data into a lollipop diagram along the length of RAD51C (Fig. 4A). A mutation cluster is apparent around the Walker A motif, which is within the best conserved region of RAD51C, such that it is largely invariant among RAD51C homologs in other organisms (SI Appendix, Fig. S1 A and B). Moreover, this region also shares 50% identity with RAD51 itself (Fig. 4A). Variants at eight residues that are conserved with RAD51 were all defective for HR, whereas variants at three residues that are not conserved with RAD51 were all proficient in HR and/or interactions with other RAD51 paralogs (SI Appendix, Fig. S4A). Another conserved residue, K131, has also been reported to be essential for RAD51C function (30, 31).
Fig. 4.
RAD51C variants in Walker A region exhibit DNA binding and ATP hydrolysis defects in the context of the BCDX2 complex. (A) A lollipop diagram of RAD51C cancer and population variants summarizes defects observed in Y2H/Y3H and HR analysis (data from Fig. 1 and 2). The three circles represent results from XRCC3 Y2H, RAD51B Y2H, and RAD51D Y3H analyses, and the diamond represents HR activity in U2OS cells. A mutation cluster is observed around the RAD51C Walker A motif, which is expanded below with an alignment to RAD51 for RAD51C amino acids 120 to 142. RAD51C variants at conserved residues show defects in all assays, whereas those in nonconserved residues do not. The Inset shows a detail of a RAD51 structure around the ATPase catalytic site (Protein Data Bank [PDB] 5NWL) with conserved residues highlighted, with numbering from RAD51C. Residues at or around the Walker A motif are colored in salmon and those from the Walker B motif are in purple. ATP and its bound magnesium ion are also shown. (B) WT RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) complexes or complexes containing the indicated RAD51C mutants were purified in the presence of ATP and MgCl2 and loaded onto a Superpose 6 size exclusion column. Fractions were run on a sodium dodecyl-sulfate polyacrylamide gel electrophoresis gel and visualized by Coomassie staining, and those containing the BCDX2 complexes used for subsequent experiments are indicated (arrows). BC (RAD51B-RAD51C) and DX2 (RAD51D-XRCC2) were run as doublets, with RAD51C and RAD51D as the upper bands in the respective doublets. Complexes with RAD51C C135Y appear less stable with distinct DX2 subcomplexes apparent in the later fractions. Protein markers are as follows: Tg, thyroglobulin; Fe, ferritin; Al, aldolase; Ov, ovalbumin. (C) ssDNA binding. ssDNA binding by the BCDX2 complexes was assessed by EMSAs using an 80mer substrate in the presence of ATP and MgCl2. The ssDNA shifted complex with C135Y runs slightly faster on the gels. (D) ATP hydrolysis. ATP hydrolysis was assessed by incubating BCDX2 complexes with ATP over time in the absence or presence of ssDNA and performing thin-layer chromatography with polyethyleneimine cellulose sheets. Pi indicates released inorganic phosphate after hydrolysis. The graph shows results from three independent experiments in the presence of ssDNA. (E) Summary of defects observed with RAD51C variants in the Walker A mutation cluster that were analyzed biochemically.
We examined the mutation cluster in the context of a RAD51 filament structure in complex with ATP-Mg2+ (32) (Fig. 4A, Inset). Within the Walker A motif itself (GX4GKT), the lysine side chain makes multiple hydrogen bonds with the β and γ phosphates of ATP, with these phosphates being involved in the coordination of the Mg2+ metal ion. The neighboring threonine interacts with the Mg2+ bound by ATP, as well as with the aspartic acid in the Walker B motif (RAD51, D222; RAD51C, D242). The glycine residue that precedes the conserved lysine in the Walker A motif flanks a loop connecting the alpha helix and a beta strand. Based on the above, RAD51C variants that harbor changes in these Walker A residues likely impair HR by adversely affecting ATP binding and/or hydrolysis (see below). Defects in nucleotide binding may also be relevant to deficiency in interactions with other RAD51 paralogs, given that in RAD51 the nucleotide-binding pocket of one RAD51 monomer is in direct contact with the ATPase domain of the next RAD51 monomer (32, 33) (SI Appendix, Fig. S4D).
Conserved residues flanking the Walker A motif also make functionally important contacts with the ATP ligand in the RAD51 structure (Fig. 4A, Inset). The glutamine following the Walker A motif interacts with a conserved arginine (RAD51, Q135:R170; RAD51C, Q133:R168) that contacts adenine in ATP, and the conserved cysteine interacts with the lysine (RAD51, C137:K133; RAD51C, C135:K131) in the Walker A motif. The downstream conserved leucine (RAD51, L140; RAD51C, L138) and valine (RAD51, V142; RAD51C, V140) likely contribute to the stability of the alpha helix. Given the conservation in this region, it seems likely that RAD51C adopts a similar conformation as RAD51, which is consistent with the AlphaFold structure prediction (SI Appendix, Fig. S4E), with ATP being sandwiched between RAD51C and an adjacent RAD51 paralog (see below).
The RAD51C Mutation Cluster Causes Defects in DNA Binding and ATP Hydrolysis by the BCDX2 Complex.
Given the importance of the RAD51C Walker A region for interactions between the RAD51 paralogs in the Y2H and Y3H analysis, we investigated the stability and activities of the BCDX2 and CX3 complexes containing mutant RAD51C by protein expression and complex purification. For BCmutDX2, all four proteins were expressed in insect cells followed by a multistep purification scheme in the presence of ATP and MgCl2 to isolate the complex, including size fractionation on a Superose 6 column (Fig. 4B and SI Appendix, Fig. S5A). BCmutDX2 complexes could be purified for each of the mutants tested (G125V, G130A, T132P, Q133K, and C135Y), and the mutant complexes fractionated at the same position as the WT complex (Fig. 4B). The presence of the three other paralogs clearly helped stabilize the RAD51C mutants in the four-member complex, as BCmut and BCmutD complexes were unstable (SI Appendix, Fig. S5B). BCDX2 complexes purified in the absence of ATP and MgCl2 were less stable and frequently dissociated, which was even more pronounced with the RAD51C mutants (SI Appendix, Fig. S5C).
BCDX2 has previously been shown to bind ssDNA and hydrolyze ATP (34). We tested the biochemical activities of the BCmutDX2 complexes. For DNA binding, electrophoretic mobility shift assays (EMSAs) were performed using an 80-nucleotide (nt) ssDNA in the presence of ATP and MgCl2, as ssDNA binding was poor in their absence (SI Appendix, Fig. S5D). Whereas the WT BCDX2 complex bound ssDNA in a concentration-dependent manner, the mutant complexes were defective in DNA binding, although to different extents (Fig. 4C and SI Appendix, Fig. S5E). Complexes containing RAD51C Q133K and G130A showed intermediate DNA binding activity, whereas those with G125V and C135Y showed little activity. DNA binding by the complex containing T132P was not detectable, as we previously reported (35). Thus, mutations in the RAD51C Walker A region impair BCDX2 ssDNA binding.
The ATPase activity of the BCDX2 complex is stimulated by ssDNA (Fig. 4D and SI Appendix, Fig. S5F), suggesting that defective DNA binding in the mutant complexes would correlate with reduced ATP hydrolysis. Consistent with this prediction, each of the mutations decreased ssDNA-dependent ATPase activity, although to various extents. G125V and T132P caused severe defects in both DNA binding and ATP hydrolysis. G130A resulted in intermediate ssDNA binding and intermediate ATPase activity. On the other hand, Q133K represented a partial separation of function, as follows: while ssDNA binding was intermediate, ATPase activity was extremely low in the mutant complex, suggesting a more specific defect in either ATP binding or hydrolysis. Since the binding partner for the Q133 carbonyl oxygen is R168, which is predicted to contact the adenine in ATP, Q133K presents a clashing charge change that could disrupt ATP binding. We extended the lollipop diagram for the RAD51C mutation cluster to incorporate these biochemical results (Fig. 4E).
RAD51 paralog complexes in other species are mediators of RAD51 nucleoprotein filament assembly that results in RPA displacement from ssDNA (17–20). Therefore, we asked whether the BCDX2 complexes would bind to RAD51. Interestingly, the WT complex failed to bind RAD51 either in the presence of ATP and MgCl2 or in the presence of the nonhydrolyzable AMP-PNP and MgCl2 (SI Appendix, Fig. S5G). Thus, we did not test the mutant complexes for RAD51 binding.
We also examined the ability of the RAD51C mutants to form a complex with XRCC3 (CX3) in the presence of ATP and MgCl2. A previous attempt to purify XRCC3 in complex with the T132P mutant was unsuccessful since the protein was insoluble (35). Therefore, to aid in solubility, RAD51C was tagged with maltose binding protein (MBP) and coexpressed with Flag-tagged XRCC3 in insect cells. A two-step affinity purification protocol was used, and in this case, CmutX3 complexes could be observed for T132P and each of the other mutants (G125V, G130A, Q133K and C135Y; SI Appendix, Fig. S5H). However, gel filtration performed on these samples demonstrated that the RAD51C mutants and XRCC3 were predominantly in the void volume, possibly as aggregates, unlike the WT protein that was eluted in later fractions together with XRCC3 (SI Appendix, Fig. S5H). In case the MBP tag was interfering with complex stability, gel filtration was also performed after cleaving the MBP tag from the T132P mutant. Even with the MBP tag removed, the mutant protein again was only evident in the void volume, while the WT protein primarily eluted with XRCC3 in later fractions (SI Appendix, Fig. S5I). Given the instability of the CmutX3 complexes, biochemical assays could not be performed.
RAD51C Variants in the Mutation Cluster Lead to Decreased RAD51 Focus Formation and Therapeutic Agent Sensitivity.
We further investigated the cellular consequences of mutations within this region. DNA damage–induced RAD51 focus formation is often used as a proxy for HR proficiency and requires several HR factors, including RAD51C (25). We tested four mutants in this region that are HR deficient in reporter assays, T121R, G125V, T132P, and C135Y, and one variant that is HR proficient, A126T. The mutants were transduced into RAD51C knockout U2OS cells, and then cells were treated with either ionizing radiation (IR), which creates DSBs and other lesions, or the crosslinking agent cisplatin. RAD51C knockout S-phase cells showed very few RAD51 foci upon treatment with either agent, whereas cells complemented with WT or A126T RAD51C exhibited abundant foci (SI Appendix, Fig. S6 A and B). However, cells expressing any of the HR-deficient variants displayed few RAD51 foci, similar to the uncomplemented cells. Thus, these mutant proteins are deficient in localizing RAD51 to sites of DNA damage.
HRD has been established to lead to cellular sensitivity to therapeutic agents including PARP inhibitors and platinum compounds (15). We performed clonogenic survival assays in RAD51C U2OS knockouts stably expressing three of the RAD51C mutants, namely, T121R, T132P, and C135Y. As with T132P, cells expressing T121R or C135Y exhibited sensitivity to both olaparib and cisplatin (SI Appendix, Fig. S6 C and D). Thus, variants that fall within conserved residues in this region consistently show the hallmarks of HRD, including sensitivity to cancer therapeutics.
RAD51C Variants Identified in Ovarian Cancer Patients by BROCA Sequencing.
BROCA is a targeted capture, massively parallel sequencing test developed to identify mutations in genes relevant to the HR pathway (39, 40). Using this approach, RAD51C variants were identified at low frequency in a multicenter study. We previously reported on a patient with a germ line T132P variant (35). Other variants in the Walker A region were also identified, namely, T121R (somatic) and Q133K (germ line in 2 cancers), as well as a variant in another RAD51-conserved residue, R312W (germ line in one cancer, somatic in another cancer) (SI Appendix, Table S5A). Although T121R is clearly a deleterious mutation in functional assays, the cancer did not exhibit LOH at RAD51C and also contained a mutation in BRCA2, such that a contribution of T121R to neoplastic initiation/progression/response in this particular patient seems unlikely. For the Q133K and R312W variants, however, three of the cancers showed evidence of LOH (tumor DNA was not available for the second Q133K patient). Each one also had TP53 mutations, although no other BROCA mutations (SI Appendix, Table S5 B and C). The family of the patient with the germ line R312W exhibited an extensive history of breast and ovarian cancer, similar to what has been previously reported for this mutation (41). The patients were treated with platinum drugs/Taxol and surgery, and remarkably, the three patients with identified LOH at RAD51C exhibited exceptional outcomes, with survival >10 y postdiagnosis, which is similar to what has been reported for the patient with the T132P variant (35).
Another RAD51C variant in the Walker A region, G130A, has been identified through Memorial Sloan Kettering clinical sequencing in a patient with invasive triple-negative breast cancer (SI Appendix, Table S5A). The variant was somatic with evidence of LOH in the cancer. The patient was treated with the neoadjuvant adriamycin/cyclophosphamide followed by carboplatin/Taxol. At the time of mastectomy, she achieved a pathologic complete response and is without evidence of disease 3 y after treatment. It remains to be seen how these (and other) deleterious RAD51C variants impact the therapeutic responses of patients.
Structural Modeling of the BCDX2 and CX3 Complexes Is Consistent with RAD51C Variant Analysis.
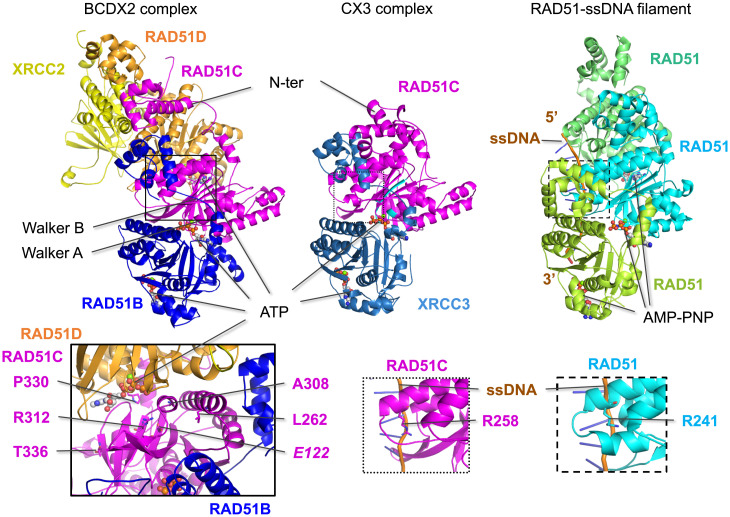
We generated models of the BCDX2 and CX3 complexes using AlphaFold2 version 1 (36–38) and evaluated them in the context of our experimental results. In the BCDX2 complex, RAD51B and RAD51D are predicted to bind on opposite sides of RAD51C, whereas XRCC2 would primarily bind RAD51D (Fig. 5), as in previous molecular analyses (summarized in ref. 16). In the CX3 complex, XRCC3 is predicted to bind RAD51C in the same manner as RAD51B. In both complexes, ATP binding can be modeled between the paralogs, as it is between RAD51 monomers, with RAD51C-bound ATP adjacent to RAD51B in the BCDX2 complex and adjacent to XRCC3 in the CX3 complex. The structural models highlight the importance of the RAD51C Walker A region, as was seen in pairwise Y2H interactions with RAD51B and XRCC3.
Fig. 5.
Predicted structures of the BCDX2 and CX3 complexes. Structural predictions of the BCDX2 and CX3 complexes were generated by AlphaFold2. RAD51C and the other RAD51 paralogs are predicted to share structural similarity to RAD51 in the ATPase domain, which contains the Walker A and Walker B motifs (highlighted in cyan for RAD51C) and to bind ATP at the subunit interfaces, similar to RAD51 binding of AMP-PNP in the nucleoprotein filament (PDB 5h1b). RAD51B and XRCC3 are predicted to bind on the same surface of RAD51C, while RAD51D is predicted to bind to the opposite side from RAD51B in the BCDX2 complex. Mutations that interfere with ATP binding are expected to impact subunit interactions and, therefore, complex formation. This is borne out in the biochemical instability of CmutX3 complexes, while BCmutDX2 complexes are more stable, possibly because of the greater number of interactions. Left Inset: Five RAD51C variants that specifically interfere with the RAD51D interaction in Y3H assays are in residues that are predicted to be close to RAD51D, with P330 positioned the closest. Among these is R312, which hydrogen bonds with E122 (italics); notably, both of these residues are conserved among all of the RAD51 paralogs. Middle and Right Insets: A key residue that contacts ssDNA in the RAD51 presynaptic filament is R241, which corresponds to R258 in RAD51C and can be modeled to bind ssDNA.
While a major interface of RAD51C with the other paralogs is at its ATPase domain, its N-terminal helical domain is predicted to interact with the N-terminal domains of the adjacent paralogs, except for XRCC2 that does not have this domain (SI Appendix, Fig. S7A). Specifically, the BCD N-terminal domains contact each other in order and also contact the adjacent RAD51 paralog’s ATPase domain (Fig. 5 and SI Appendix, Fig. S7B). Similarly, in CX3, the RAD51C N-terminal domain interacts with the XRCC3 N-terminal domain, although it is configured differently than in the BCDX2 complex. These predicted N-terminal domain interactions are a major distinction from RAD51 filaments, in which the N-terminal domains are relatively isolated from each other (SI Appendix, Fig. S7B).
The different conformations of the RAD51C N-terminal domain predicted in the BCDX2 and CX3 complexes may be relevant as to why RAD51D does not bind to the CX3 complex (34). To further explore this idea, we asked how similar the N-terminal domains are in the BC subcomplex to the CX3 complex. In the absence of RAD51D and XRCC2, the N-terminal domain of RAD51C is more extended than in the CX3 complex (SI Appendix, Fig. S7C), suggesting that packing of the RAD51C and XRCC3 N-terminal domains is specific to their interactions (see also legend, SI Appendix, Fig. S7C).
Many of the variants we examined in the RAD51C N-terminal domain are in residues that are predicted to be solvent exposed (e.g., P21, K26, P43). This prediction is consistent with the proficiency of the variants in Y2H/Y3H interactions and in HR (Fig. 2B). On the other hand, L27 is predicted to be buried within the N-terminal domain. Mutation to proline may sufficiently impinge on the structure to affect HR while not affecting interactions with the N-terminal domains of the other paralogs.
The predicted structural interactions would explain the frequent loss of all three interactions in Y2H and Y3H assays for many of the variants. A single RAD51C variant could directly affect both RAD51B and XRCC3 binding, and the loss of RAD51B binding would impair RAD51D binding in the BCDX2 complex. This may explain why the loss of the RAD51D interaction correlates the best with HR levels, as it can capture defects in binding RAD51B and XRCC3, as well as RAD51D specifically. Furthermore, the intertwined interactions between RAD51C and the other RAD51 paralogs in the predicted BCDX2 structure could account for the biochemical stability of mutant complexes compared to the instability of the mutant CX3 complexes.
While RAD51C Walker A mutants have global interaction defects, a specific perturbation of RAD51D binding in Y3H assays was observed with some of the variants. For example, P330 is structurally positioned closest to RAD51D (and away from RAD51B and XRCC3) (Fig. 5, Left Inset) and when mutated to histidine (P330H) substantially impairs HR. Another of these residues, R312, is more buried within the RAD51C structure itself and is predicted to form a salt bridge with E122, which is located just upstream of the Walker A motif, which in turn is predicted to form a salt bridge with N284, as does the cognate residues in RAD51. Both R312 and E122 are part of the small number of residues that are identical among all of the paralogs, and N284 is also highly conserved (SI Appendix, Fig. S7A). Thus, we would predict that like R312W, mutation of either E122 or N284 would result in defective HR.
A cryo-electron microscopy structure of a RAD51 presynaptic filament was previously reported with RAD51 in complex with ssDNA and the nonhydrolyzable ATP analog AMP-PNP (Fig. 5) (42). One of the major side chain contacts with the phosphate backbone of DNA is residue R241. The cognate residue in RAD51C is R258, which can be modeled to bind ssDNA in a similar manner to RAD51 R241 (Fig. 5, Insets). This residue is also conserved in RAD51B and XRCC3 (SI Appendix, Fig. S7). The R258H variant found in Fanconi anemia patients has minimal defects in interactions with the other three RAD51 paralogs but is hypomorphic for HR activity (Fig. 1 C and F). Thus, we speculate that RAD51C (and the other paralogs) binds ssDNA similarly to RAD51 in the presynaptic filament.
Discussion
We present here the most comprehensive analysis yet of missense variants in the tumor suppressor RAD51C, with a focus on variants identified in breast and ovarian cancers. We also present structural predictions for the two major RAD51C-containing complexes that provide coherent working models for understanding effects of variants on RAD51C functional activity. Several conclusions can be drawn from this study. While the majority of variants were deemed deleterious by three in silico prediction tools (PolyPhen-2, SIFT, and PROVEAN), only about half were actually deficient in the canonical process involving RAD51C, namely, HR. Thus, the prediction tools were limited in their ability to accurately infer function. Conservation of the variant residue with the cognate residue in RAD51 is as strong or a better predictor of HR activity and its proxy RAD51D interaction as the in silico tools because few of the variants in nonconserved residues were defective in either activity. Thus, incorporating RAD51 conservation into predictive tools, in addition to RAD51C conservation itself, is expected to make them more robust. Moreover, the importance of the RAD51-conserved residues provides insight into how RAD51C interacts with the other RAD51 paralogs to function in HR.
More recently, ensemble prediction tools have been developed, including Rare Exome Variant Ensemble Learner (REVEL), which incorporates the 3 in silico tools we used as well as 15 other assessors of variant pathogenicity (43). When applied to the RAD51C variants, we found that REVEL performed better than the individual in silico tools. For the most part, variants proficient in the HR and RAD51D interaction were separated in the REVEL scoring from those deficient in these two activities, although the REVEL scores for the latter were spread over a wide range (SI Appendix, Fig. S8 A and B).
Another finding from our study was that HR activity associated completely with the ability of variants to support the proliferation of RAD51C conditional MCF10A cells. Essentiality assays have already been applied to BRCA1 for VUS calling using saturation gene editing in functionally important domains (44). The value to this approach is the scale in that 4,000 single-nucleotide variants in BRCA1 could be assigned using the survival of the haploid cell line HAP1 as the readout. Our study, therefore, predicts that essentiality assays would also be valuable for RAD51C VUS predictions.
We found that results from yeast interaction assays correlated well with HR activity, specifically interactions with known RAD51C binding partners, the RAD51 paralogs RAD51B, RAD51D, and XRCC3. RAD51D interaction correlated the best with HR activity (r = 0.85 for RAD51C-AD fusions), such that RAD51D interaction could provide an alternative screening approach to direct HR assays for RAD51C VUS analysis. Like all functional assessments, interaction assays may result in false negatives, if interactions are maintained, but HR activity itself is defective. However, we observed only one variant, namely, L27P, that gave that result, indicating that L27P has an HR defect downstream of RAD51 paralog complex formation.
The most striking finding from the yeast assays was that several RAD51C variants were defective for interactions with the three other RAD51 paralogs, identifying the Walker A motif and proximal residues as a critical RAD51C region. The structural models predict ATP binding at the interface of RAD51C interactions with other RAD51 paralogs, similar to interactions between RAD51 monomers in filaments. RAD51C variants in this conserved region with RAD51 show consistent HR deficiency, accompanied by impaired DNA damage–induced RAD51 foci formation and sensitivity to the therapeutic agents cisplatin and olaparib. The observed sensitivity of these RAD51C Walker A VUSs is explained molecularly by impaired CX3 complex formation and impaired biochemical activity within the BCDX2 complex. Thus, the cluster of mutations within the RAD51C Walker A region forms a hot spot for RAD51C functional impairment. This region also has the most deleterious REVEL scores (SI Appendix, Fig. S8C and Table S6A).
While none of the five RAD51C Walker A mutants we tested was able to form a stable CX3 complex, each was able to form a BCDX2 complex, likely through stabilization by the other three RAD51 paralogs. ATPase activity was reduced in each mutant BCDX2 complex, in keeping with the mutation location within conserved residues in or near the Walker A box. Defects in ssDNA binding were also observed, which may more specifically indicate defects in ATP binding (45). ATP hydrolysis may promote the dissociation from ssDNA, which remains to be tested, as with other RAD51 family members. Interestingly, the variants in this region were not equivalent to each other. In biochemical assays, T132P had the most severe defects, while G130A showed intermediate ssDNA binding and ATPase activity and also a modicum of HR activity (0.32). This distinction likely reflects, at least partially, the amino acid substitution in the variant; proline is expected to disrupt the alpha helix in which T132 is situated, whereas the alanine side chain is expected to be less disruptive at the beta turn in which G130 is located. Thus, although both residues are within the catalytic center of RAD51C, it would be expected that the particular missense mutation would impact the functionality of the variant, although in this case, both variants show HRD.
Our analysis here focused on RAD51C variant protein interactions and HR function. However, RAD51C has additional functions during replication fork protection and restart (46, 47). To date, the loss of RAD51C replication–associated functions alone is not predictive of cancer risk or therapeutic response. Therefore, the impact of the cancer variants on replication was not assessed here but is an important consideration for future analysis.
We sought to classify the RAD51C variants we analyzed as pathogenic or benign based upon the criteria from the American College of Medical Genetics and Genomics (ACMG) standards (48) (SI Appendix, Table S6B). The ACMG guidelines take into account several criteria for variant calling, including computational predictions, cosegregation of the variant with affected individuals, and functionality. Therefore, our functional assays (viability, HR, RAD51 paralog interactions) help inform the recommendation as benign or pathogenic or the likelihood of either. Assessments from established prediction tools that rely on RAD51C conservation were considered together with RAD51 conservation. Previously reported results were also incorporated, including variant segregation within affected families (15 variants) and RAD51C variant functionality (11 variants). Published functional studies typically involved some of the same assays we used, although the advantage from our study is the large number of variants assessed side by side. Based upon the ACMG standards, we classified the 56 RAD51C variants we tested (SI Appendix, Table S6). For previously analyzed variants, our recommendations are concordant. For newly analyzed variants, our recommendations are that 17 additional variants be classified as likely pathogenic, including the Walker A region variants G130A, Q133K, and C135Y for which extensive biochemical analysis was performed. Our recommendations extended to several variants in the less well-conserved N terminus for which previous studies were limited. Six variants beginning with Q11R and extending to P43S were considered to have uncertain significance by ACMG criteria, at least in part due to the low frequency in the general population and scores on prediction tools. However, because they were fully functional for HR, we consider that it is more likely that they are benign variants and suggest giving functionality more weight in the recommendations. On the other hand, we cannot rule out that RAD51C has some other function besides HR that is important for tumor suppression.
We recently reported the long-term survival (>10 y) of an ovarian cancer patient harboring the RAD51C T132P variant (35). Here, we report the long-term survival of three other ovarian cancer patients with deleterious RAD51C variants (Q133K, R312W). A breast cancer patient with another deleterious variant, G130A, has also been identified but is just 3 y past diagnosis. Although the durable patient responses could be due to successful surgical removal of the cancer, chemosensitization of the cancer by these variants could also have played an important or even a more critical role.
In addition to sensitizing the cancer to initial therapy, certain RAD51C variants may also impact the likelihood of developing secondary somatic reversion mutations associated with therapeutic resistance. Cancers associated with BRCA1 or BRCA2 mutations often develop chemoresistance through secondary reversion mutations that restore the open reading frame. Similarly, an ovarian cancer patient with a RAD51C mutation also developed PARP inhibitor resistance (7). The original mutation led to a truncation at a nonconserved residue (R193X); several different reversion alleles restored the reading frame, leading to HR proficiency and therapy resistance. By contrast, reversion events might be less tolerated in critical residues in a protein. Because the missense variants G130A, T132P, and Q133K are in the Walker A region of RAD51C, we hypothesize that each of these mutations would be highly constrained in its ability to undergo reversion to an alternative allele, such that tumors carrying the mutations would maintain a robust and durable chemotherapeutic response. Although not in the Walker A region, R312W is also a residue that is conserved with RAD51, as are many other RAD51C residues that are critical for its function; they may be similarly constrained in their ability to revert, requiring restoration of the original residue or a change to a very limited number of residues. Patient follow-up will be necessary to determine the impact of these RAD51C mutations, and likely other RAD51 paralog mutations, on long-term outcome.
Materials and Methods
Additional information is available in the SI Appendix, Materials and Methods.
Mutational Predictive Analysis.
Polymorphism Phenotyping version 2 (PolyPhen-2) (23), Sorting Intolerant From Tolerant (SIFT; RRID:SCR_012813) (24), and Protein Variation Effect Analyzer (PROVEAN; RRID:SCR_002182) (22, 49) were used to predict the functional effects of the amino acid substitution for each variant.
Cellular Analyses.
Clonogenic survival assays and HR analysis were performed using MCF10A RAD51C conditional cells (25), which contain an integrated DR-GFP reporter (50). RAD51C mutant U2OS cells with an integrated SCR reporter were cultured as described previously for clonogenic survival and HR assays (25). RAD51C variant expression in U2OS cells was detected by Western blot.
Y2H and Y3H Analysis.
Y2Hs and Y3Hs were performed as described in ref. 51 except that the pGAD, pGBD, and pRS-ADH-416 plasmids were cotransformed into the YPJ69-4a yeast strain. Western blotting was performed with extracts from yeast expressing RAD51C variants from the pGAD-C1 (AD expression vector) and XRCC3 expressed in the pGBD-C1 (BD expression vector).
BCDX2 and CX3 Complex Biochemistry.
Hi5 insect cells were infected with baculoviruses expressing RAD51B-His, RAD51C variants, RAD51D, and XRCC2-Flag (BCDX2 complex) and MBP-RAD51C variants and XRCC3-Flag (CX3 complex). For DNA binding, 1 nM of 5′ 32P-labeled 80-nt ssDNA (20) was incubated with the indicated concentration of the purified BCDX2 complex. For ATPase assays, BCDX2 complexes (2.0 μM) were incubated in the absence or presence of ssDNA (20 μM nucleotides, phiX174 virion) with 0.05 mM [ϒ-32P] ATP. The level of ATP hydrolysis was determined by layer chromatography on PEI cellulose F sheets (Millipore, 105579) in 0.375 M potassium phosphate (pH 3.5).
Structural Modeling of the BCDX2 and CX3 Complexes.
Sequences of human RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3 were retrieved from the UniProt database (RRID:SCR_004426 (52)) and used as input in the mmseqs2 homology search program (53) to generate a multiple sequence alignment (MSA) against the UniRef30 clustered database (38) for each RAD51 paralog. Special care was taken in selecting only orthologs from the MSA of each paralog. The models are available in ModelArchive (www.modelarchive.org) with the accession codes ma-a54ps (BCDX2) and ma-52hi1 (CX3).
Patient Mutations.
This study was approved by the Mayo Clinic and MSK Institutional Review Boards. All patients provided written informed consent for genomic study of their ovarian and breast cancers, respectively, including DNA sequencing.
Supplementary Material
Acknowledgments
We thank David Schild for XRCC3, Jun Huang and Paul Russell for plasmids, Weibin Wang for baculoviruses, and Pei Xin Lim for discussions. Funding is acknowledged from NIH F31 ES027321(M.R.S.); P50 CA136393 (S.H.K.); SU2C-AACR-DT16-15 (E.M.S., S.H.K., and M.J.); Department of Defense (DOD) OC120506 and OC120312 (E.M.S); French National League against Cancer (équipe labellisée) and the French National Cancer Institute (M.M.); R01 ES007061 and R35 CA241801 (P.S.); MSK Functional Genomics Initiative, P30 CA008748, and R35 CA253174 (M.J.); and R01 ES031796, DOD BC201356, SU2C-AACR-IRG-02-16, and V Foundation for Cancer Research V2014-019 (K.A.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202727119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
References
- 1.Chen C. C., Feng W., Lim P. X., Kass E. M., Jasin M., Homology-directed repair and the role of BRCA1, BRCA2, and related proteins in genome integrity and cancer. Annu. Rev. Cancer Biol. 2, 313–336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pennington K. P., et al. , Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin. Cancer Res. 20, 764–775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tung N., et al. , Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J. Clin. Oncol. 34, 1460–1468 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pritchard C. C., et al. , Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N. Engl. J. Med. 375, 443–453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor M. J., Targeting the DNA damage response in cancer. Mol. Cell 60, 547–560 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Lord C. J., Ashworth A., PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondrashova O., et al. ; AOCS Study Group, Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 7, 984–998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ollier M., et al. , DNA repair genes implicated in triple negative familial non-BRCA1/2 breast cancer predisposition. Am. J. Cancer Res. 5, 2113–2126 (2015). [PMC free article] [PubMed] [Google Scholar]
- 9.Maxwell K. N., et al. , BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat. Commun. 8, 319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meindl A., et al. , Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet. 42, 410–414 (2010). [DOI] [PubMed] [Google Scholar]
- 13.LaDuca H., et al. , A clinical guide to hereditary cancer panel testing: Evaluation of gene-specific cancer associations and sensitivity of genetic testing criteria in a cohort of 165,000 high-risk patients. Genet. Med. 22, 407–415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scully R., Panday A., Elango R., Willis N. A., DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan M. R., Bernstein K. A., RAD-ical new insights into RAD51 regulation. Genes (Basel) 9, ••• (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prakash R., Zhang Y., Feng W., Jasin M., Homologous recombination and human health: The roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 7, a016600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sung P., Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11, 1111–1121 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Belan O., et al. , Single-molecule analysis reveals cooperative stimulation of Rad51 filament nucleation and growth by mediator proteins. Mol. Cell 81, 1058–1073.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy U., et al. , The Rad51 paralog complex Rad55-Rad57 acts as a molecular chaperone during homologous recombination. Mol. Cell 81, 1043–1057.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaines W. A., et al. , Promotion of presynaptic filament assembly by the ensemble of S. cerevisiae Rad51 paralogues with Rad52. Nat. Commun. 6, 7834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang M. T., et al. , Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 34, 155–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi Y., Chan A. P., PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei I. A., et al. , A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sim N. L., et al. , SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40, W452-7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcin E. B., et al. , Differential requirements for the RAD51 paralogs in genome repair and maintenance in human cells. PLoS Genet. 15, e1008355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaz F., et al. , Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 42, 406–409 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Schild D., Lio Y. C., Collins D. W., Tsomondo T., Chen D. J., Evidence for simultaneous protein interactions between human Rad51 paralogs. J. Biol. Chem. 275, 16443–16449 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Baldock R. A., et al. , RAD51D splice variants and cancer-associated mutations reveal XRCC2 interaction to be critical for homologous recombination. DNA Repair (Amst.) 76, 99–107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang M. T., et al. , Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 8, 174–183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.French C. A., Tambini C. E., Thacker J., Identification of functional domains in the RAD51L2 (RAD51C) protein and its requirement for gene conversion. J. Biol. Chem. 278, 45445–45450 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Nagaraju G., Odate S., Xie A., Scully R., Differential regulation of short- and long-tract gene conversion between sister chromatids by Rad51C. Mol. Cell. Biol. 26, 8075–8086 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brouwer I., et al. , Two distinct conformational states define the interaction of human RAD51-ATP with single-stranded DNA. EMBO J. 37, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conway A. B., et al. , Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 11, 791–796 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Masson J. Y., et al. , Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 15, 3296–3307 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan M. R., et al. , Long-term survival of an ovarian cancer patient harboring a RAD51C missense mutation. Cold Spring Harb. Mol. Case Stud. 7, a006083 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans R., et al. , Protein complex prediction with AlphaFold-Multimer. bioRxiv [Preprint] (2021). 10.1101/2021.10.04.463034. [DOI]
- 38.Mirdita M., et al. , ColabFold:making protein folding accessible to all. Nat. Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed]
- 39.Walsh T., et al. , Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 107, 12629–12633 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norquist B. M., et al. , Mutations in homologous recombination genes and outcomes in ovarian carcinoma patients in GOG 218: An NRG oncology/gynecologic oncology group study. Clin. Cancer Res. 24, 777–783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gayarre J., et al. , Characterisation of the novel deleterious RAD51C p.Arg312Trp variant and prioritisation criteria for functional analysis of RAD51C missense changes. Br. J. Cancer 117, 1048–1062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J., et al. , Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange. Nat. Struct. Mol. Biol. 24, 40–46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ioannidis N. M., et al. , REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 99, 877–885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Findlay G. M., et al. , Accurate classification of BRCA1 variants with saturation genome editing. Nature 562, 217–222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menetski J. P., Kowalczykowski S. C., Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 181, 281–295 (1985). [DOI] [PubMed] [Google Scholar]
- 46.Somyajit K., Saxena S., Babu S., Mishra A., Nagaraju G., Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 43, 9835–9855 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berti M., et al. , Sequential role of RAD51 paralog complexes in replication fork remodeling and restart. Nat. Commun. 11, 3531 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richards S., et al. ; ACMG Laboratory Quality Assurance Committee, Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi Y., Sims G. E., Murphy S., Miller J. R., Chan A. P., Predicting the functional effect of amino acid substitutions and indels. PLoS One 7, e46688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng W., Jasin M., BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat. Commun. 8, 525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McClendon T. B., Sullivan M. R., Bernstein K. A., Yanowitz J. L., Promotion of Homologous Recombination by SWS-1 in Complex with RAD-51 Paralogs in Caenorhabditis elegans. Genetics 203, 133–145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.UniProt Consortium, UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinegger M., Söding J., MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.