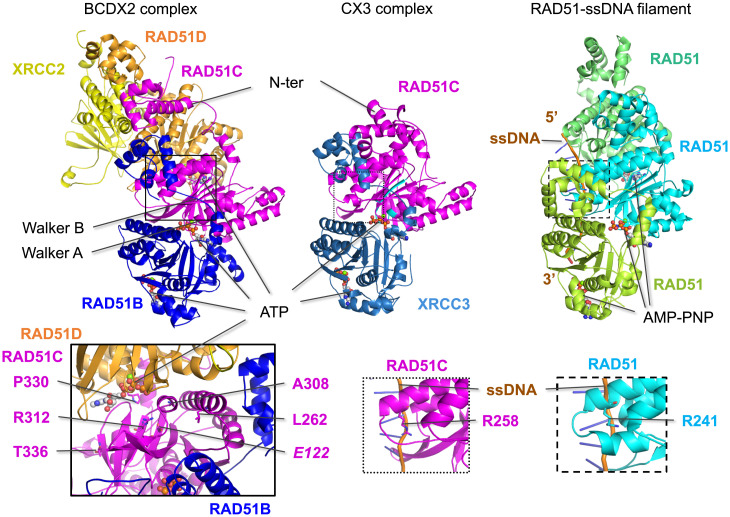
Fig. 5.
Predicted structures of the BCDX2 and CX3 complexes. Structural predictions of the BCDX2 and CX3 complexes were generated by AlphaFold2. RAD51C and the other RAD51 paralogs are predicted to share structural similarity to RAD51 in the ATPase domain, which contains the Walker A and Walker B motifs (highlighted in cyan for RAD51C) and to bind ATP at the subunit interfaces, similar to RAD51 binding of AMP-PNP in the nucleoprotein filament (PDB 5h1b). RAD51B and XRCC3 are predicted to bind on the same surface of RAD51C, while RAD51D is predicted to bind to the opposite side from RAD51B in the BCDX2 complex. Mutations that interfere with ATP binding are expected to impact subunit interactions and, therefore, complex formation. This is borne out in the biochemical instability of CmutX3 complexes, while BCmutDX2 complexes are more stable, possibly because of the greater number of interactions. Left Inset: Five RAD51C variants that specifically interfere with the RAD51D interaction in Y3H assays are in residues that are predicted to be close to RAD51D, with P330 positioned the closest. Among these is R312, which hydrogen bonds with E122 (italics); notably, both of these residues are conserved among all of the RAD51 paralogs. Middle and Right Insets: A key residue that contacts ssDNA in the RAD51 presynaptic filament is R241, which corresponds to R258 in RAD51C and can be modeled to bind ssDNA.