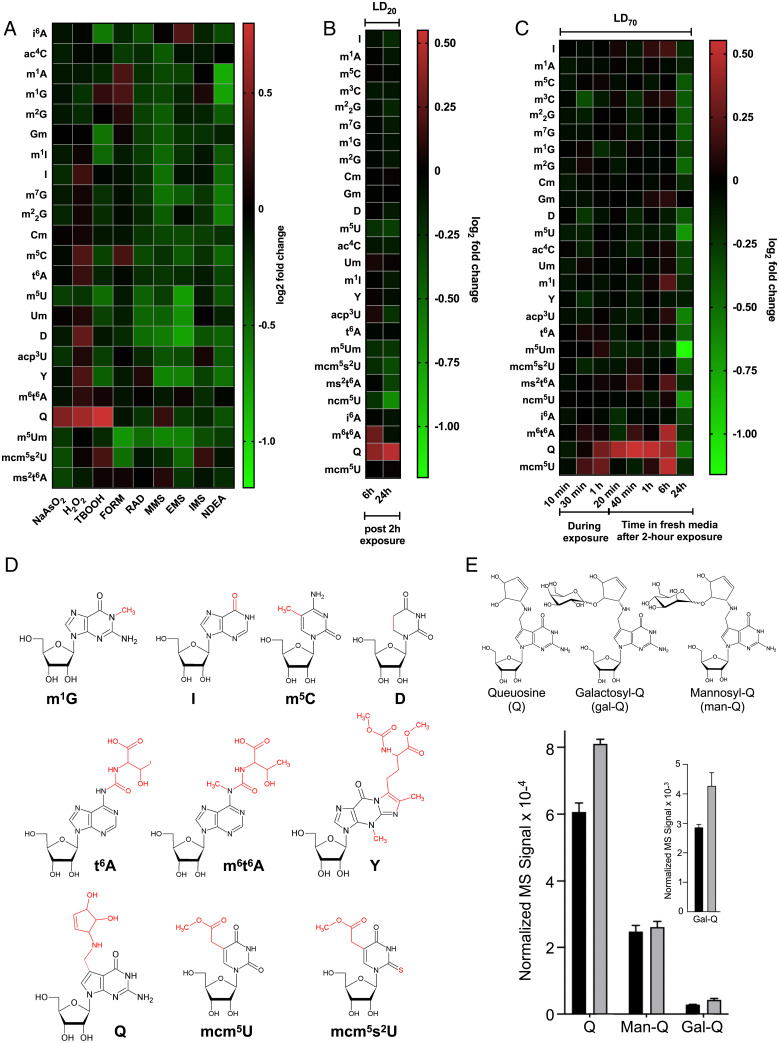
Fig. 1.
Epitranscriptomic reprogramming identifies Q and other tRNA modifications as signature markers of oxidative stress and NaAsO2 exposure. (A) tRNA modification reprogramming in HepG2 cells exposed to LD20 doses of oxidizing and alkylating agents for 2 h, with RNA harvested 24 h postexposure. (B and C) Epitranscriptomic response of HepG2 cells to (B) a 2 h exposure to an LD20 dose of NaAsO2 followed by 6 h and 24 h postexposure incubation and (C) a LD70 dose at various times postexposure. (D) Chemical structures and abbreviations of the modifications affected by oxidative stressors and NaAsO2 exposure in the present studies. (E) Q, Gal-Q, and Man-Q were quantified by LC-MS in tRNA extracted from HepG2 cells exposed to an LD20 dose of arsenite for 6 h. Black bars: unexposed; gray bars: arsenite-exposed. Inset shows a magnification of data for Gal-Q on the 10−3 scale. Data represent mean ± SD for three experimental replicates. See data in Dataset S1.