Fig. 6.
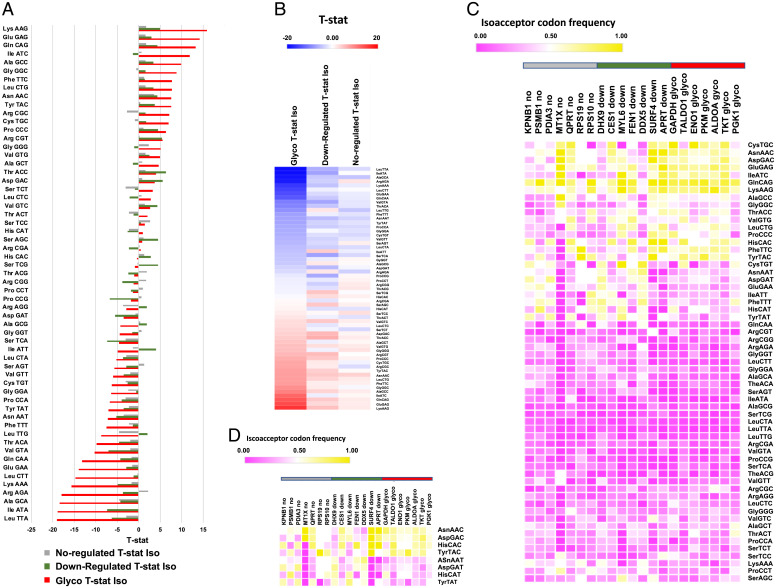
Among genes for proteins altered by arsenite exposure, glycolytic genes have distinct codon usage patterns relative to genes for other proteins identified by proteomics. Codon usage comparison of the genes specific to seven up-regulated glycolytic (glyco) proteins, seven random down-regulated (down) proteins, and seven random unchanged proteins (not-regulated; no) identified in our proteomics study. Isoacceptor T-stat values for codons specific to the three groups of gene sequences in (A) bar graph and (B) heat map view. Heat maps describing isoacceptor codon frequencies for each of seven gene sequences defining the three groups of proteins for (C) all possible codons and (D) the eight Q-decoded codons. For (C) and (D), the horizontal gray, green, and red bars highlight the no, down, and glyco genes, respectively, matching the color coding used in (A).