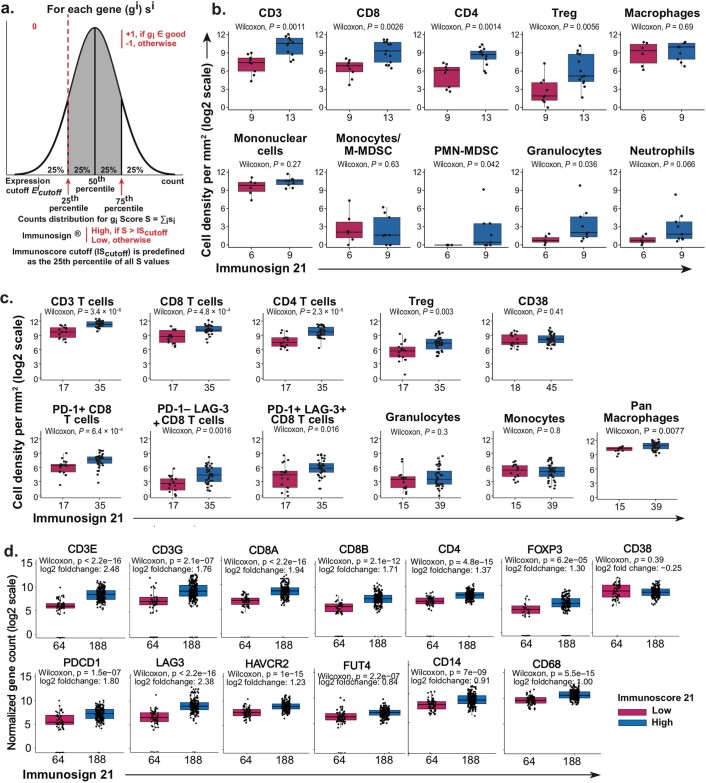
Extended Data Fig. 6. Correlative studies of Immunosign 21 score with cell subsets across 3 independent datasets.
a, Definition of the Immunosign 21 score cutoff. The Immunosign 21 scoring function is an algorithm derived from the Immunoscore algorithm, is independent of clinical outcome, and is predefined as the 25th percentile of the observed scores among samples. b-d, Correlation of Immunosign 21 (low versus high) with cell subsets in 3 independent datasets (Two-sided Wilcoxon test). b,c, Immunosign 21 correlations with cell subset densities analysed by Immunoscore TCE/TCE+ panels in b, ZUMA-1 pretreatment tumour biopsy specimens from third line, r/r DLBCL patients (subset 1, n = 22 for T-cell subsets; n = 15 for myeloid subsets) and c, DLBCL biopsy specimens at diagnosis from treatment-naïve patients (n = 67). d, Immunosign 21 correlations with normalised gene expression of T-cells or of myeloid cells analysed by IO360 NanoString panel in pretreatment tumour biopsy specimens from second line DLBCL patients (n = 252). The number of samples per group is indicated on each panel. DLBCL, diffuse large B-cell lymphoma; FOXP3, forkhead box P3; LAG-3, lymphocyte-activation gene 3; M-MDSC, monocytic myeloid-derived suppressor cell; PD-1, programmed cell death protein 1; PMN-MDSC, polymorphonuclear myeloid-derived suppressor cell; SC, Suppressive cell; TCE, T-cell Exhaustion; TIM-3, T-cell immunoglobulin and mucin domain 3; Treg, regulatory T-cell.