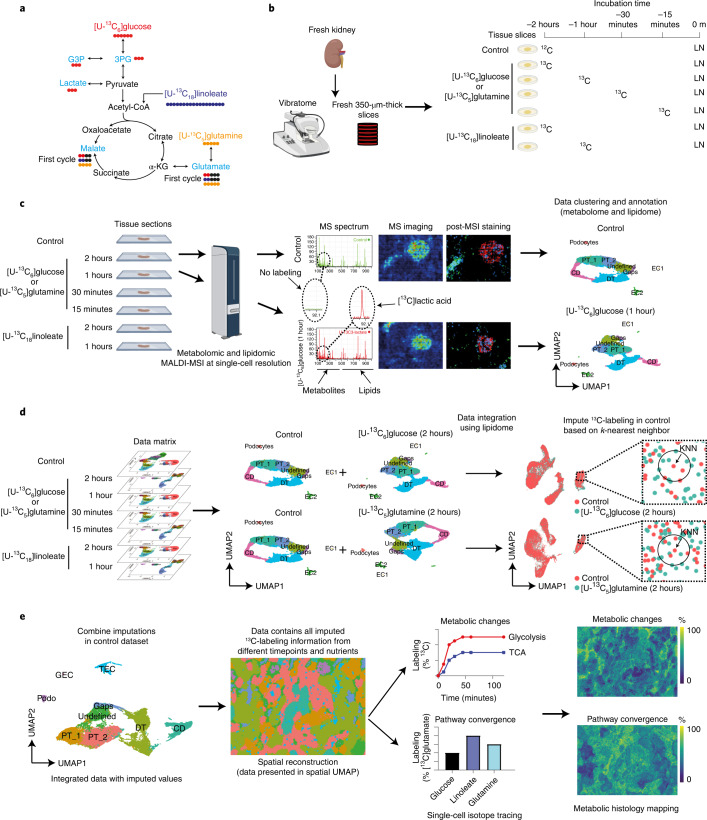
Fig. 1. Workflow of cell-type-specific dynamic metabolic measurements and analysis.
a, Overview of the traced isotopes in the primary carbon metabolism. The contributions of U-13C-labeled nutrients ([U-13C6]glucose, [U-13C18]linoleate, and [U-13C5]glutamine) to glycolytic and TCA intermediates (light blue) were traced. b, Fresh mouse kidney tissue was cut into 350-µm-thick slices using a vibratome. For 13C isotope tracing in tissue culture, different 13C-labeled nutrients were added to a well-defined medium described in the Methods section at different timepoints. Samples were quenched using liquid nitrogen (LN). c, The metabolome and lipidome were measured in all samples using MALDI-MSI at high spatial resolution (5 × 5 µm2 pixel size). MALDI-MSI data were preprocessed and transferred into a data matrix. Cell types were identified on the basis of lipid profiles and IF staining after MALDI-MSI. Images in b and c were created using Biorender. d, Cell-type-specific (phospho)lipid data were used to characterize various cell types. The lipid data were used for anchor-based data integration of the ‘control’ data matrix with data matrices of sections from 13C-isotope tracing measurements. We used KNN analysis to impute the molecular information contained in the 13C-labeling timecourse data matrices into the control data matrix. e, Establishment of the imputed dataset, in which each pixel contains all added 13C-labeling information from each timepoint and labeled nutrient. Dynamic metabolic calculations were performed on single pixels, including metabolic rates and pathway convergence. To visualize the heterogeneity in tissue metabolic dynamics, a series of pseudoimages, which were generated from calculated values, were created by tracing pixel coordinates back to the original spatial information from the MALDI-MSI analysis. α-KG, alpha-ketoglutaric acid; DT, distal tubules; CD, collecting ducts; PT, proximal tubules; EC, endothelial cells; GEC, glomerular endothelial cells; TEC, peritubular endothelial cells.