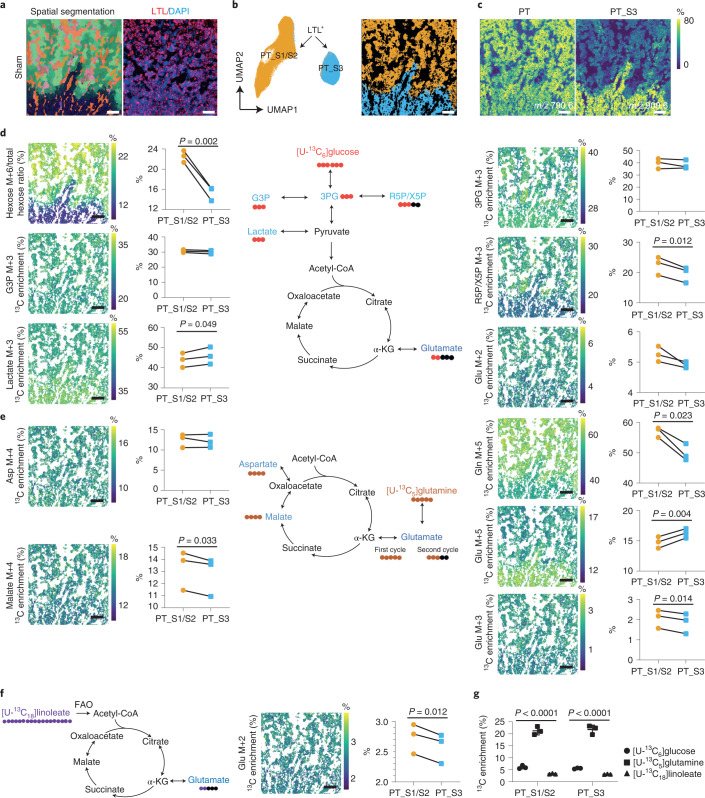
Fig. 3. Dynamic metabolic measurements on sham kidney PT cells.
a, Left, representative molecular histology of cortical and outer stripe of outer medulla areas of sham kidney (n = 3), generated from three-dimensional UMAP analysis on the basis of lipid profiles recorded by MALDI-MSI (5 × 5 μm2 pixel size). Right, LTL immunofluorescent staining on post-MALDI-MSI tissue (red). b, 2D UMAP plot and representative spatial segmentation showing lipid heterogeneity between LTL-positive proximal tubular cells from the cortical and outer stripe of outer medulla areas of sham kidneys (n = 3). c, Representative images showing lipid species distribution in the cortical and outer stripe of outer medulla areas of sham kidney (n = 3), as recorded by MALDI-MSI (5 × 5 μm2 pixel size). d, Dynamic metabolic measurements using [U-13C6]glucose on PT cells of sham kidneys. e, Dynamic metabolic measurements using [U-13C5]glutamine on PT cells of sham kidneys. f, Dynamic metabolic measurements using [U-13C18]linoleate on PT cells of sham kidneys. Images showing the average 13C enrichment of isotopologues on tissue. Graphs showing the comparison of the average 13C enrichment of isotopologues between PT S1/S2 and PT S3. The average 13C enrichment (area under curve (AUC) normalized to total time) of isotopologues were derived from [U-13C6]glucose or [U-13C5]glutamine measured at different timepoints (0, 15, 30, 60, and 120 minutes) or [U-13C18]linoleate measured at different timepoints (0, 60, and 120 minutes). Charts showing the traced isotopes and their derived isotopologues. Two-tailed paired t-test was performed. All scale bars, 200 µm. g, Direct carbon contribution of different nutrients to glutamate at the 2-hour timepoint, as measured from the glutamate isotopologues M+2, M+3, and M+5. One-way analysis of variance (ANOVA) was performed (n = 3). Glc, glucose; 3PG, 3-phosphoglycerate; G3P, glycerol-3-phosphate; R5P/X5P, ribulose-5-phosphate/xylulose-5-phosphate; α-KG, alpha-ketoglutaric acid; Glu, glutamate; Gln, Glutamine; Asp, aspartate.