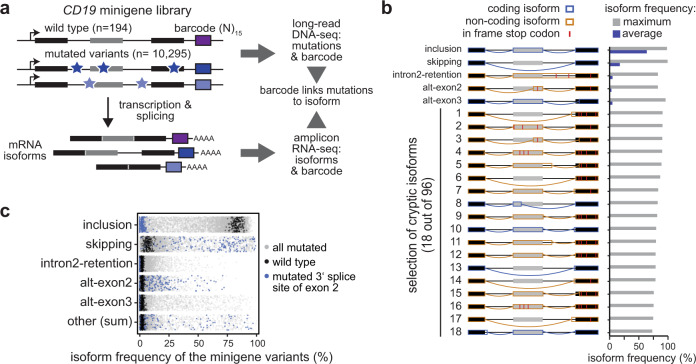
Fig. 2. High-throughput mutagenesis identifies splicing-affecting mutations and cryptic isoforms in the CD19 minigene.
a High-throughput detection of splicing-affecting mutations and cryptic isoforms. Mutagenic PCR creates mutated minigene variants (top) that upon transfection into NALM-6 cells give rise to alternatively spliced transcripts (bottom). Mutations (stars) and corresponding splicing products are characterised by DNA and RNA sequencing, respectively, and linked by a unique 15-nt barcode sequence in each minigene (coloured boxes). Black and grey boxes depict constitutive and alternative exons, respectively. b A large number of CD19 splice isoforms arise in the minigene library. CD19 splice isoforms with the highest maximal isoform frequency across all 9321 minigene variants. Schematic representation (left) of 5 major and 18 cryptic isoforms depicts exons 1–3 (boxes) and introns (horizontal lines) with splice junctions for each isoform (arches). Colour indicates coding potential (blue, coding; orange, non-coding). In-frame stop codons are indicated by red lines. Bar graph (right) shows average and maximal isoform frequency across all minigenes. Cryptic isoforms are sorted by maximal isoform frequency (Supplementary Data 2). c Inclusion isoform dominates in WT minigenes, whereas mutated variants show broad spread in all major isoforms. Frequencies of five major isoforms in replicate 1 for all wild type (black; n = 195) and mutated (grey; n = 9476) minigenes in the library. Minigene variants harbouring a mutation in the 3′ splice site of exon 2 (n = 174) are highlighted in blue. “Other” refers to the sum of 96 cryptic isoforms.