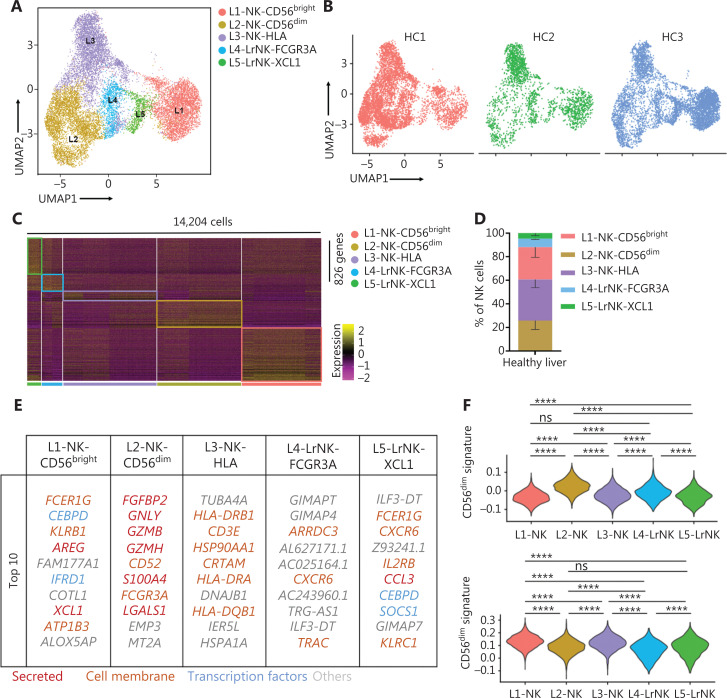
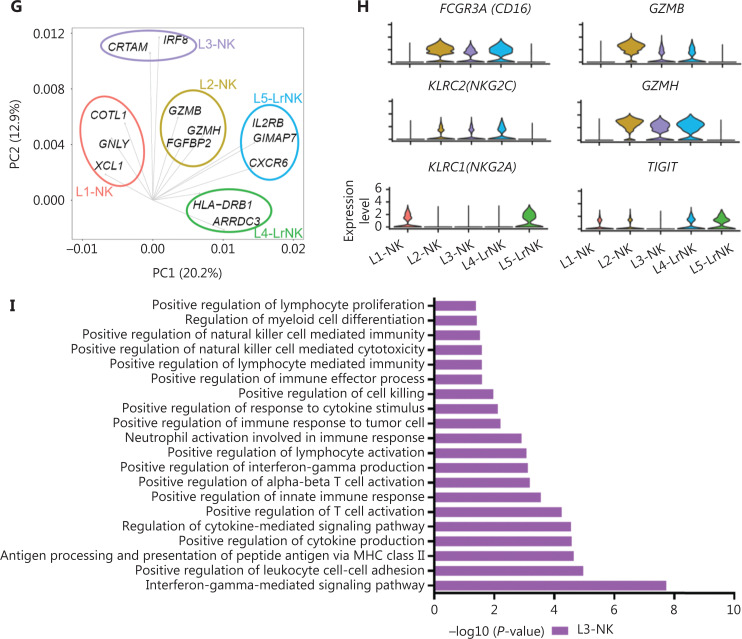
Figure 3.
High-throughput scRNA-seq identifies 5 NK cell subsets in the healthy human liver. (A) UMAP projection of NK cells from an integrated analysis of NK cells from 3 healthy control donors, showing the formation of 5 main subsets identified according to the gene expression characteristics of each subset. (B) UMAP plot of human liver NK cells from healthy livers, colored by donor (HC1: 6,234 cells; HC2: 2,124 cells; and HC3: 5,846 cells). (C) Heatmap of the 826 genes assessed with a Wilcoxon rank sum test separating the 14,204 NK cells into 5 main subsets, shown in different colors. Squares identify specific transcriptomic signatures of the distinct cell subsets. (D) The proportions for the 5 subsets defined among NK cells from the 3 healthy control donors across all liver NK cells. (E) Top 10 genes significantly differentially expressed (Wilcoxon rank sum test) among the 5 liver NK cell subsets. Genes are ranked by log2 fold-change. (F) Module scores of CD56dim and CD56bright gene expression programs defined by Hanna et al.43. Violin plots representing the distribution of module scores for CD56dim (top) CD56bright (bottom) for each liver NK cell subset (Kruskal-Wallis ANOVA followed by Dunn’s multiple comparisons test). Nonsignificant (ns) P > 0.05; ****P < 0.0001. (G) PCA for the 5 liver NK cell subsets’ driving genes. (H) Violin plots comparing the expression of selected NK cell markers in this scRNA-seq dataset. The violin represents the probability density at each value. (I) Selected Gene Ontology terms using genes upregulated (log2 fold-change > 0.25) within the L3 subset with an adjusted P < 0.05.