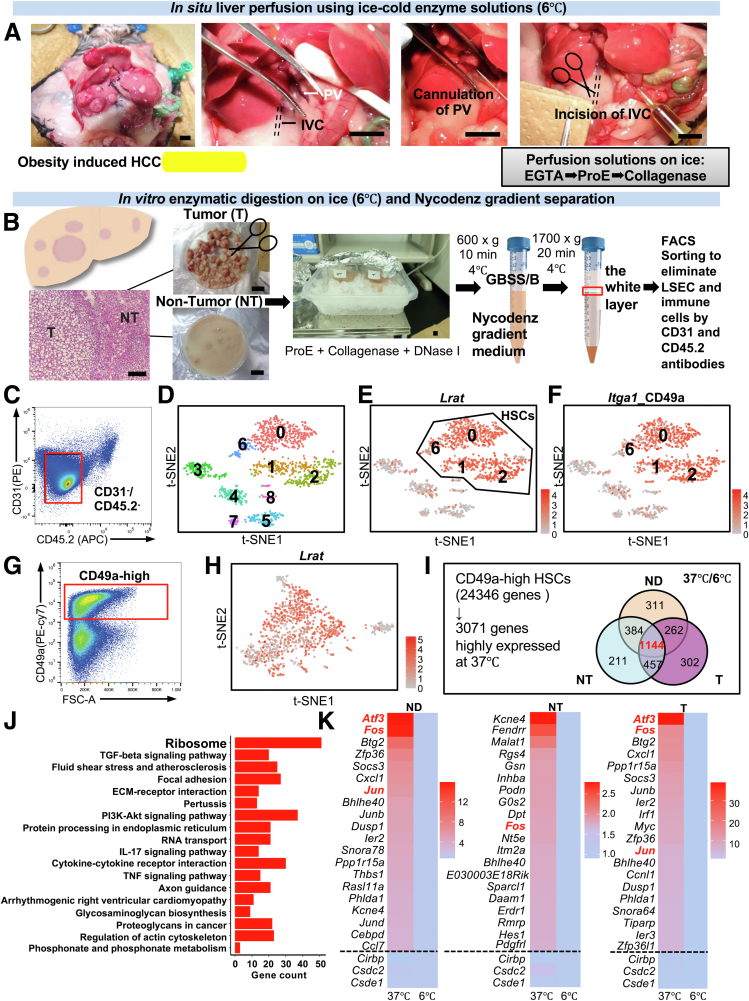
Figure 1.
Cold perfusion method for isolating HSCs minimising heat-induced artefact gene expression. CD49a is a useful HSC marker. A, The cannulation of liver via portal vein and incision of the inferior vena cava were performed in obesity-induced HCC mice (scale bars, 6 mm). The liver was perfused sequentially with EGTA, Pronase E (ProE), and collagenase. B, The histology of steatotic liver tumor in mice (hematoxylin and eosin staining, scale bar, 200 μm). In vitro enzymatic digestion and Nycodenz gradient separation. C, Flow cytometry plots for sorting CD31-/CD45.2- cells from livers of ND- and HFD-induced HCC mice. n = 6, 35 w, (3 ND mice; 3 HCC mice). The average mouse body weight (BW): ND, 36.3 g (range, 35–38 g); HCC, 58.4 g (range, 55–61 g). D, t-SNE plots of cell clusters based on single-cell transcriptomes. E, Unsupervised clustering analysis identified HSC clusters based on Lrat expression. F, Unsupervised clustering analysis of Itga1 among CD31-/CD45.2- cells. G, Flow cytometry plots for sorting CD49a-high cells among CD31-/CD45.2- cells from livers of HFD-induced HCC mice. n = 6, 35 w, (3 ND mice; 3 HCC mice). The average mouse body weight (BW): ND, 31.5 g (range, 30–33 g); HFD, 53.2 g (range, 45–57 g). H, Consistent unsupervised clustering analysis of Lrat using CD49a-high cells among CD31-/CD45.2- cells. I, Bulk RNA-sequencing analysis of CD49a-high HSCs of ND, NT, and T tissues. n = 12, 35 w, (6 ND mice; 6 HCC mice). The average mouse body weight (BW): ND, 31.7 g (range, 28–34 g); HCC, 57.6 g (range, 51–64 g). J, Pathway enrichment analysis of the 1144 common highly expressed genes using Enrichr. K, Heatmap of the top 20 commonly overexpressed genes (above the dotted line) at 37 °C among the ND, NT, and T datasets. Heat map of the cold shock genes (below the dotted line) among the ND, NT, and T datasets.