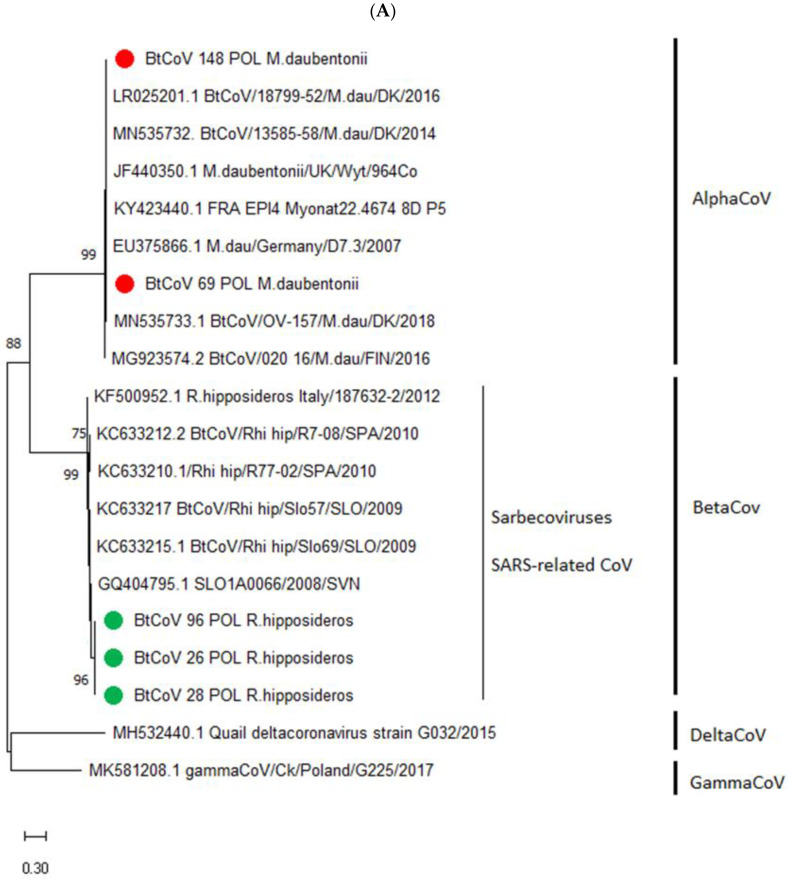
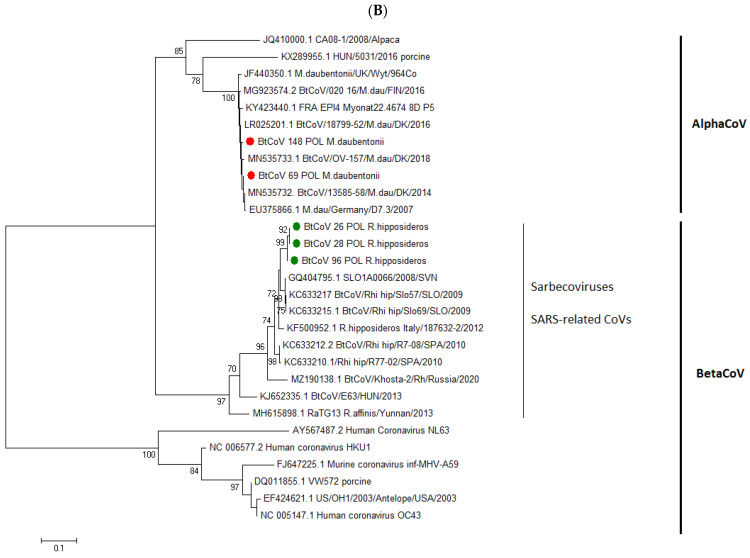
Figure 2.
Phylogenetic tree based on the 379 bp-long nucleotide fragment of the RNA-dependent RNA polymerase gene of BtCoVs selected as the most homologous (A) and BtCoVs detected in Poland and the other distinct CoVs (B), generated using the neighbor-joining method with the Kimura2-parameter model and MEGA 5 software. Green circles indicate SARS-related CoVs detected in horseshoe bats in Poland, whereas red circles refer to alphacoronaviruses detected in Myotis bats in Poland. Bootstrap values (1000 replicates) over 70% indicating significant support for the tree topology are shown next to the branches. Sequences of delta- and gamma CoVs isolated from poultry in Poland were used as the outgroup in Figure 2A.