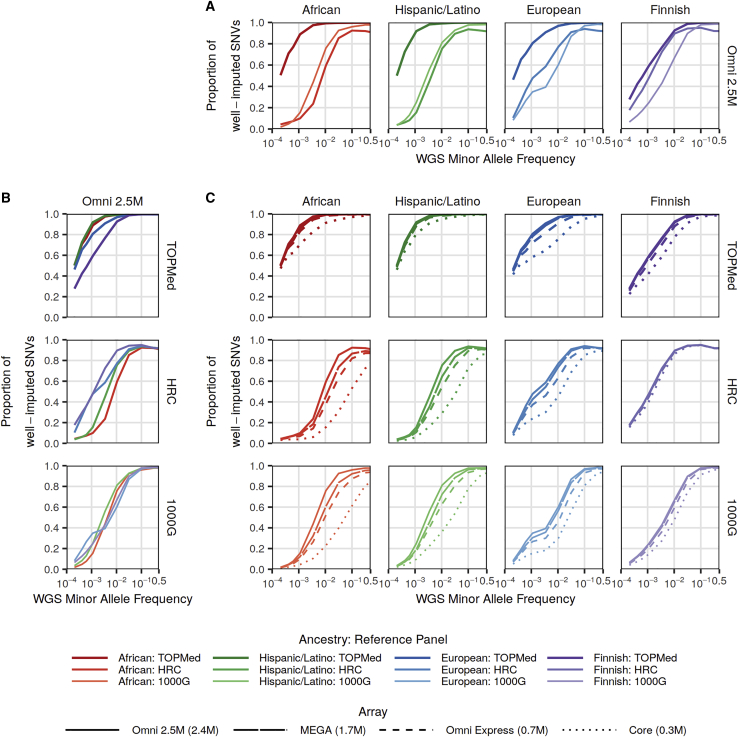
Figure 1.
Proportion of well-imputed (r2 > 0.8) bi-allelic SNVs by reference panel, study ancestry, and genotyping array
The proportion of sequenced variants that are well-imputed (r2 > 0.8) with the TOPMed, HRC, and 1000G imputation reference panels.
(A) Comparison across the reference panels using the Illumina Omni 2.5M array.
(B) Comparison across the four studies using the Illumina Omni 2.5M array.
(C) Comparison across four Illumina genotyping arrays: Omni 2.5M, MEGA, Omni Express, and Core by ancestry (columns) and imputation reference panels (rows). In all plots, the x axes show minor-allele frequency (MAF) calculated separately by study. Sequenced bi-allelic SNVs not present in reference panels were assigned r2 = 0. Bi-allelic SNVs were then aggregated by MAF bins of width 0.00025 MAF for MAF between 0.0002 and 0.002 and of size 0.001 MAF for MAF > 0.002; those plotted here correspond to singletons, doubletons, and tripletons in each study, as well as those with mean MAF closest to the values 0.001, 0.0032, 0.01, 0.032, 0.1, 0.32, and 0.5.