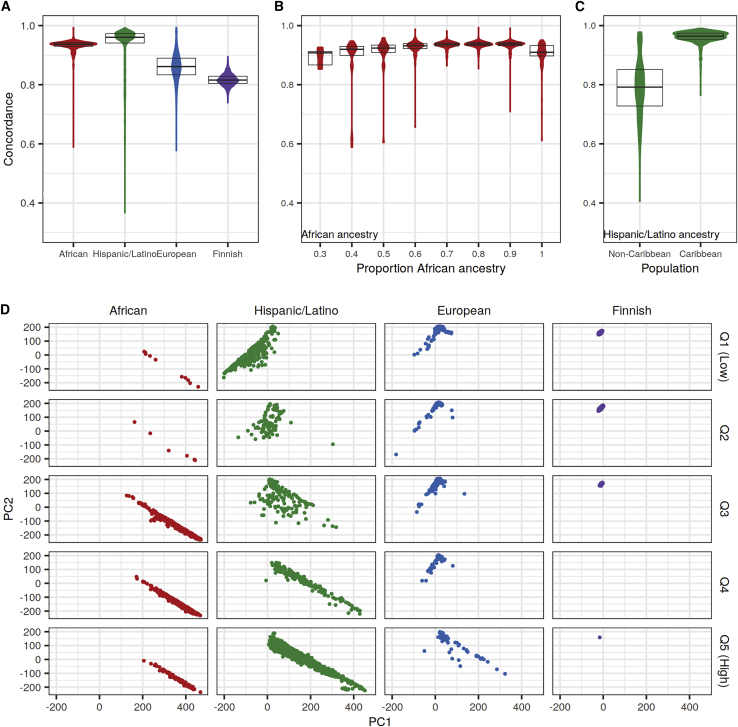
Figure 2.
Heterozygous genotype concordance rates for rare variants by ancestry with TOPMed panel imputation
Heterozygous concordance rates were calculated between sequenced and TOPMed-imputed genotypes for rare (MAF < 0.5%, calculated separately in each study) bi-allelic SNVs with the Omni 2.5M array.
(A) Distribution of concordance rates in each of the four studies. Boxplots correspond to 25th, 50th, and 75th percentiles.
(B) Distribution of concordance rates by bins of estimated proportion of African ancestry in the admixed African study.
(C) Distribution of concordance rates in Caribbean and non-Caribbean populations in the Hispanic/Latino study.
(D) Principal-component analysis (PCA) by genotype concordance quintile and ancestry. PCA was performed by projecting onto the Human Genome Diversity Project reference samples. Genotype concordance quintiles were calculated across all four studies and correspond to concordance rates of 0.37–0.82 (Q1), 0.82–0.86 (Q2), 0.86–0.93 (Q3), 0.93–0.95 (Q4), and 0.95–0.99 (Q5). Points are colored by ancestry.