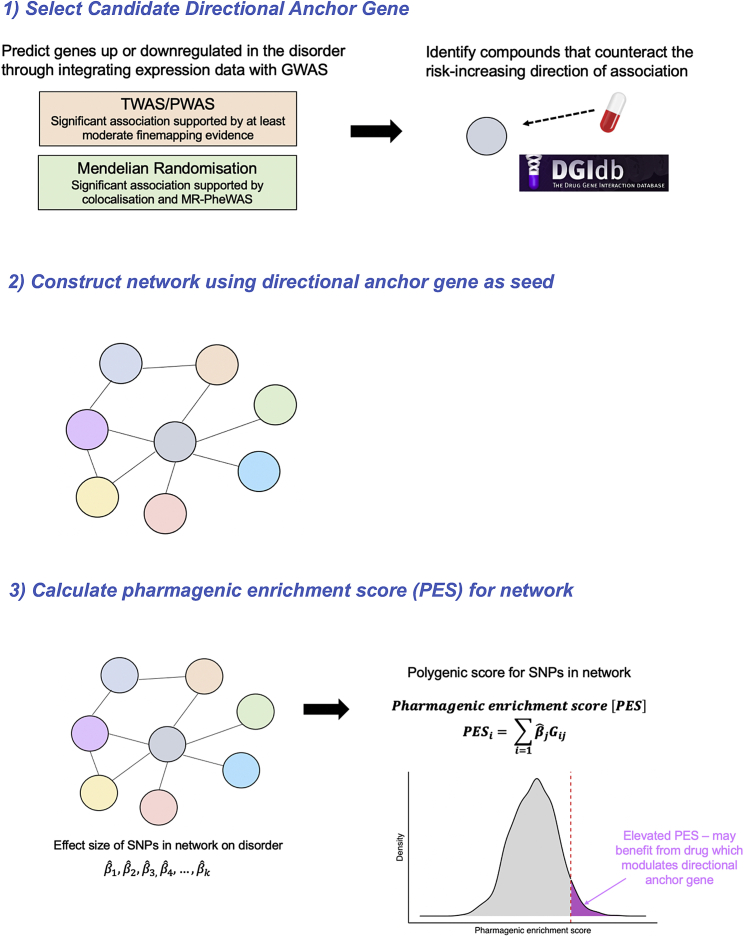
Figure 1.
Overview schematic of the integration of candidate directional anchor genes with pharmagenic enrichment scores
Directional anchor genes are genes targeted by an approved compound, in what is genetically predicted (through integration with expression data) to decrease the risk of the disorder or modulate the trait in a clinically useful manner. For instance, if increased expression of a gene is associated with a disorder through a transcriptome or proteome-wide association study (TWAS/PWAS) or Mendelian randomization (MR) using quantitative trait loci as instrumental variables, then an antagonist of said gene may be a repurposing opportunity. Directional anchor genes then act as seed genes to define a network of other genes that interact with them. SNPs mapped to this network are then utilized to construct a pharmagenic enrichment score (PES) for the network. In the case of a binary disease phenotype, the interpretation of the PES would be that individuals with an elevated score relative to an appropriate population reference may benefit from a compound which modulates the directional anchor gene. The hypothesis underlying this is that these individuals will have genetic risk that impacts upstream or downstream processes relating to the directional anchor, as well as the anchor gene itself, which could be addressed by the repurposing candidate in question.