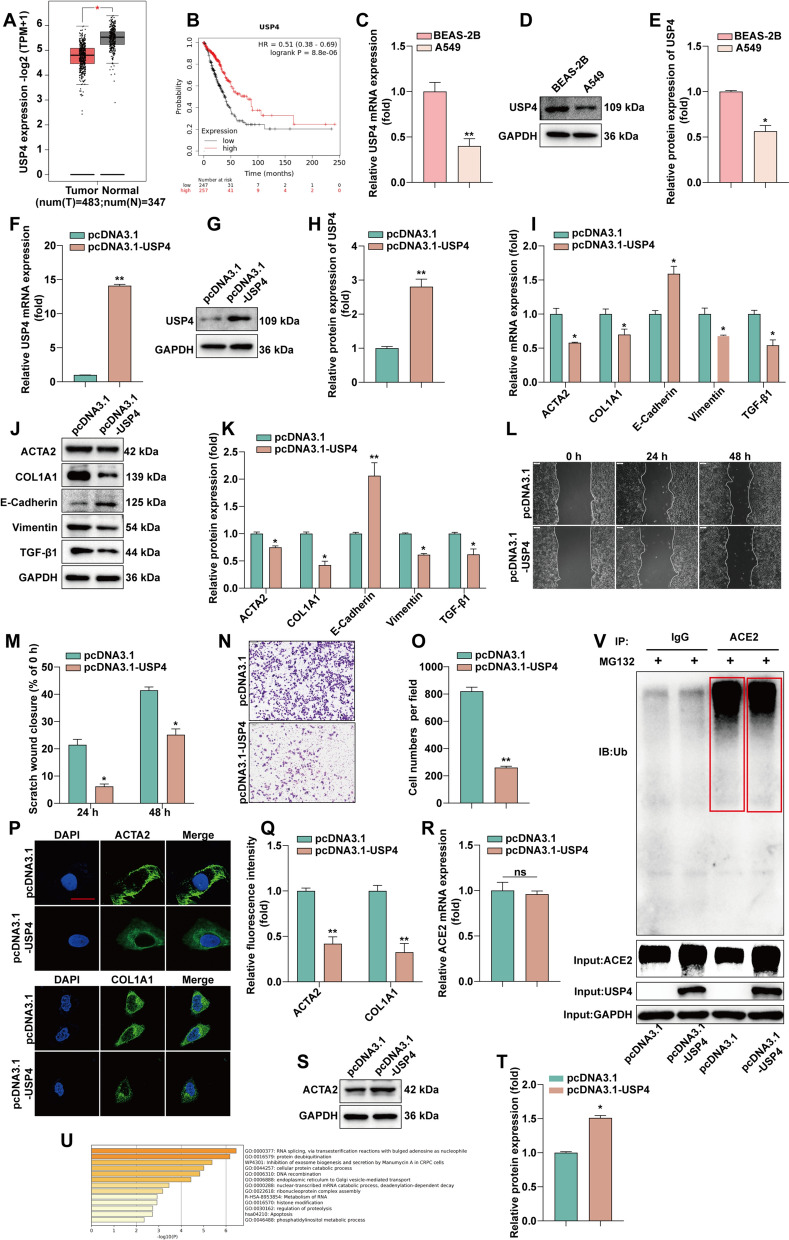
Fig. 5.
USP4 deubiquitination maintains ACE2 stability and thus inhibits EMT and fibrosis in LUAD. A Analysis of USP4 expression in LUAD and normal lung tissues using the GEPIA database. B Analysis of USP4 expression in relation to OS of LUAD patients using Kaplan–Meier plotter. C Detection of USP4 mRNA expression in BEAS-2B and A549 by RT-qPCR. D, E Detection of USP4 protein expression in BEAS-2B and A549 by WB and statistical quantification by ImageJ software. F RT-qPCR detection of USP4 mRNA expression levels after overexpression of USP4. G, H WB detection of USP4 protein expression levels after overexpression of USP4 and statistical quantification by ImageJ software. I Overexpression of USP4 followed by RT-qPCR for EMT and fibrosis marker expression. J, K The protein expressions of TGF-β1, E-cadherin, vimentin, ACTA2 and COL1A1 were detected by WB after overexpression of USP4. Statistical quantification by ImageJ software. L, M After overexpression of USP4, cell migration was monitored by wound scratch assay. N, O After the expression of USP4, cell invasion was determined by invasion assay. And counted statistically by ImageJ. P, Q After overexpression of USP4, an immunofluorescence assay was performed to detect the expression of fibrosis-associated proteins. Statistical data are shown. R Detection of ACE2 mRNA expression by RT-qPCR after overexpression of USP4. S, T Detection of ACE2 protein expression by WB after overexpression of USP4 and quantified by ImageJ software statistics. U Functional enrichment analysis of USP4 by Metascap website. V Analysis of ubiquitinated degradation of ACE2 after overexpression of USP4 by Co-IP assay