Fig. 2.
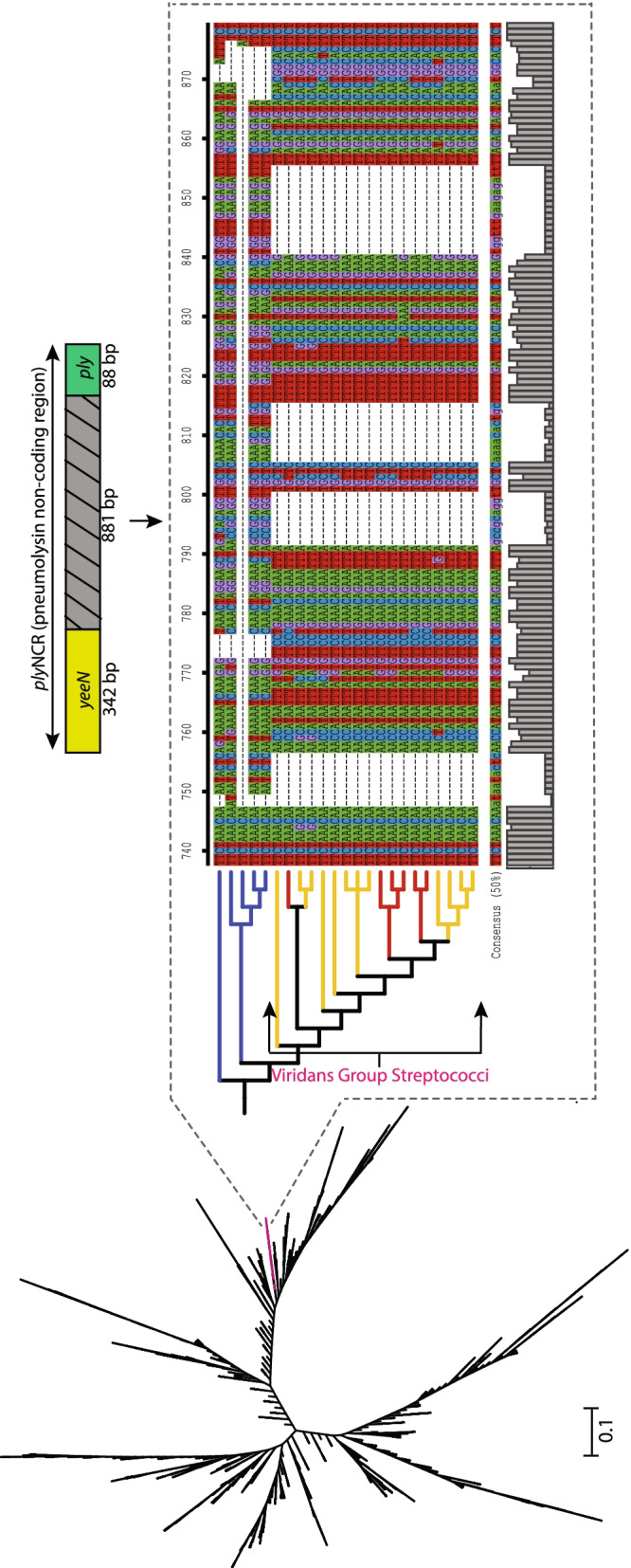
Maximum likelihood phylogeny of the plyNCR marker among strains of the Streptococcus mitis group. The ML tree was inferred using the generalized time reversible (GTR) + gamma model with FastTree and is unrooted with unscaled branches. A zoomed-in subtree (dashed rectangle) shows a cladistic clustering and a portion of the sequence alignment from five pneumococci (in blue) and 18 viridans group streptococci (VGS; S. mitis and S. pseudopneumoniae in red and yellow, respectively) with the latter having characteristic deletions. Above the subtree, the plyNCR is illustrated as a non-coding region flanked by partial sections of the conserved genes, yeeN (hypothetical regulatory protein) and ply (pneumolysin)
