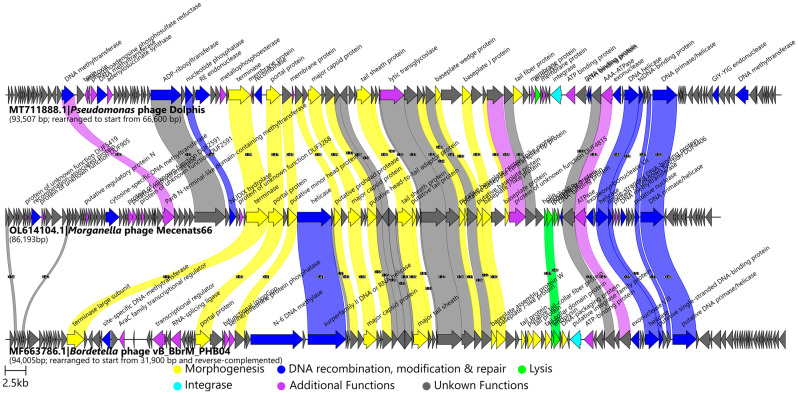
Figure 3.
Pairwise genome organization and encoded protein comparison of Morganella phage Mecenats66 (middle) to Pseudomonas phage Dolphis (upper) and to Bordetella phage vB_BbrM_PHB04 (lower). Genomes are drawn to scale, and the genomes of Dolphis and PHB04 were reordered as indicated to ensure collinearity with Mecenats66; the scale bar indicates 2500 base pairs. Arrows representing open reading frames point in the direction of the transcription and are color-coded based on the function of their putative product according to the legend. Slanted labels above the arrows indicate the predicted function for the given ORF putative product in the case it had a function assigned (original annotations were retained for Dolphis and PHB04; predicted Mecenats66 ORF123 crossing the genome termini junction is not shown). Ribbons connect homologous proteins of phages and are colored according to the predicted functional group of the respective Mecenats66 ORF product. Ribbon labels indicate the pairwise similarity between the connected ORF products (for products having ≥30% identity). Comparisons were carried out using clinker (v.0.0.23.; [65]).