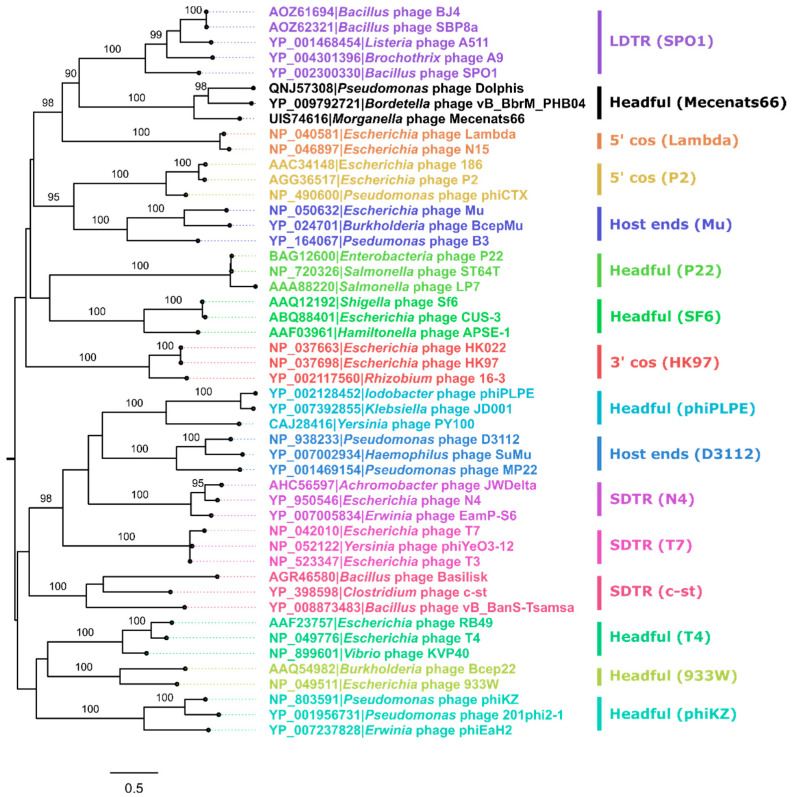
Figure 4.
The maximum-likelihood tree of terminase/terminase large subunit (TerL) amino acid sequences for the prediction of the phage Mecenats66 packaging strategy. Input alignment had 48 sequences with 1539 columns, 1482 distinct patterns, 996 parsimony-informative, 346 singleton sites, and 197 constant sites. The tree was built using VT + F + R4 as the best-fit substitution model. Near zero-length branches were collapsed into polytomies. The tree shown is midpoint rooted. Tip labels are colored based on the distinct packaging strategies the phages seen in the tree employ (experimentally verified for most of the phages represented in the tree [34]). The percentage of replicate trees in which the associated sequences clustered together in the ultrafast bootstrap (UFBoot; 1000 replicates) is shown next to the branches for branches having a UFBoot support higher or equal to 95% (except for the MRCA node of the SPO1 type and Mecenats66 type TerL/terminase sequence clades for which a UFBoot of 90% is also shown). The tree is drawn to scale, and branch lengths represent the number of amino acid substitutions per site. Tip labels correspond to the phages from which the respective terminase/TerL amino acid sequences were derived and are in the format of “Protein accession|Phage”. Colored bars next to the labels indicate evolutionary distinct TerL clades, which correspond to different packaging strategies (LDTR stands for long direct terminal repeats, SDTR—short direct terminal repeats).