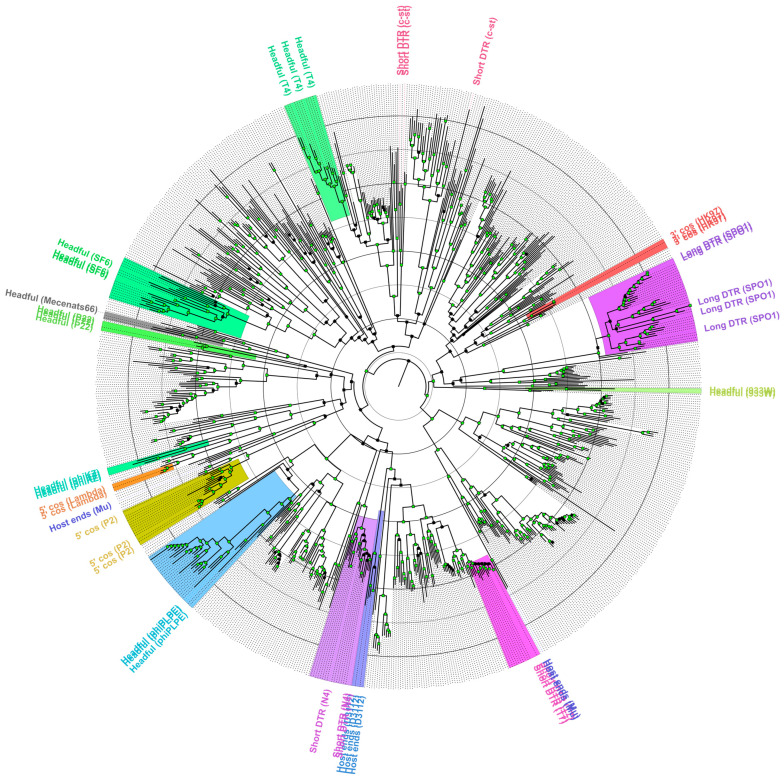
Figure 7.
The maximum-likelihood tree of representative terminase/terminase large subunit (TerL) amino acid sequence diversity. Input alignment had 793 sequences with 6470 columns, 4725 distinct patterns, 3011 parsimony-informative, 1119 singleton sites, and 2340 constant sites. The tree was built using Blosum62 + R10 as the best-fit substitution model. Near zero-length branches were collapsed into polytomies. The tree shown is midpoint rooted. Tip labels are provided for phages that had their packaging strategy/genome termini type verified experimentally and are colored based on the distinct packaging strategies phages within the presumed same packaging strategy clades employ (sequences from the extended “core” dataset have their respective packaging strategies indicated at the corresponding tips [34]; Figure 4) The clade consisting of the terminase sequence from Mecenats66, Dolphis, and PHB04 is highlighted in grey. LDTR stands for long direct terminal repeats, SDTR for short direct terminal repeats. Distal nodes of branches having UFBoot support higher or equal to 95% are indicated by green squares. The tree is drawn to scale, branch lengths represent the number of amino acid substitutions per site, and the radial circles are spaced 0.5 amino acid substitutions per site apart.