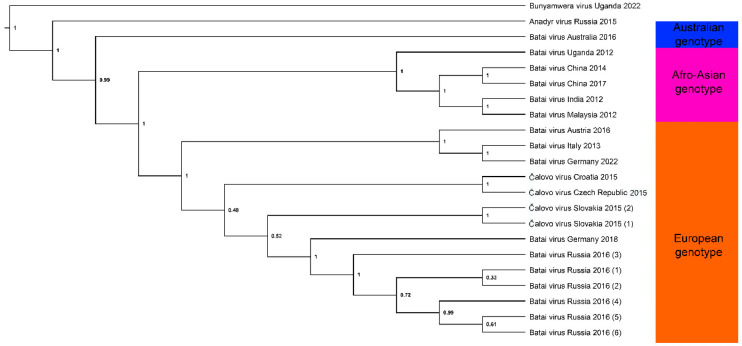
Figure 1.
Bayesian phylogenetic analysis of BATV virus, based on a 6000 bp fragment of the L-gene from 22 sequences obtained from GenBank, with BUNV used as an outgroup. Node labels show posterior probability, and three geographically distinct genotypes are highlighted. Details of isolates used are shown in Supplementary Table S1. Isolates classed as ‘Čalovo virus’ on GenBank have retained this nomenclature in our phylogenetic analyses. Repeat detections from the same year and county were differentiated using numbers in parentheses. Briefly, sequences obtained from GenBank were aligned using MAFFT v7.471 and imported into BEAST v1.10.4 where the GTR+I substitution model and 10,000,000 Markov chain Monte Carlo generations were used to create the Bayesian phylogeny. Log files were analyzed in Tracer v1.7.1, and the resulting tree was edited in FigTree v1.4.4.