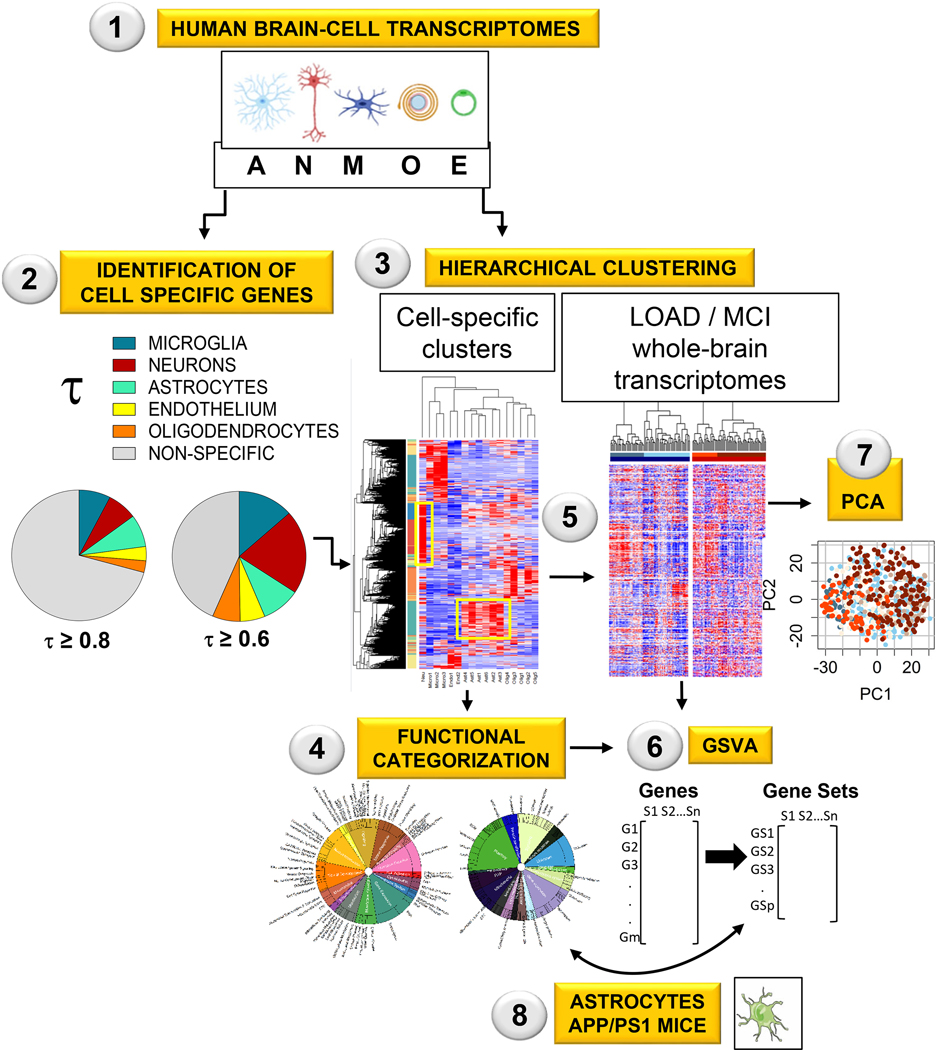
Figure 1. Workflow for the identification of altered astrocytic functions in AD and MCI (See Methods for details).
Step 1–3: Genes from human-brain cortex were classified as cell-specific or enriched using RNAseq data from cells isolated from aged healthy human brain cortices (Step 1), the univariate cell-type enrichment τ scoring (Step 2), and hierarchical clustering (Step 3).
Step 4: The astrocytic and neuronal gene clusters were manually categorized into functions and subfunctions.
Step 5: Control, AD, and MCI whole-brain transcriptomes from the MtSINAI, MAYO and ROSMAP databases were hierarchically sorted according to the cell-type specific clusters.
Step 6–7: Alterations of astrocytic and neuronal functional categories in AD and MCI groups vs Controls were statistically established using gene set variation analysis (GSVA, Step 6), and principal component analysis (PCA, Step 7).
Step 8: To compare patients with mice, the published transcriptome of APP/PS1 astrocytes was reanalyzed using our functional categorization and GSVA. The figure in Step 1 was created using cell art adapted from Servier Medical Art (https://smart.servier.com/).