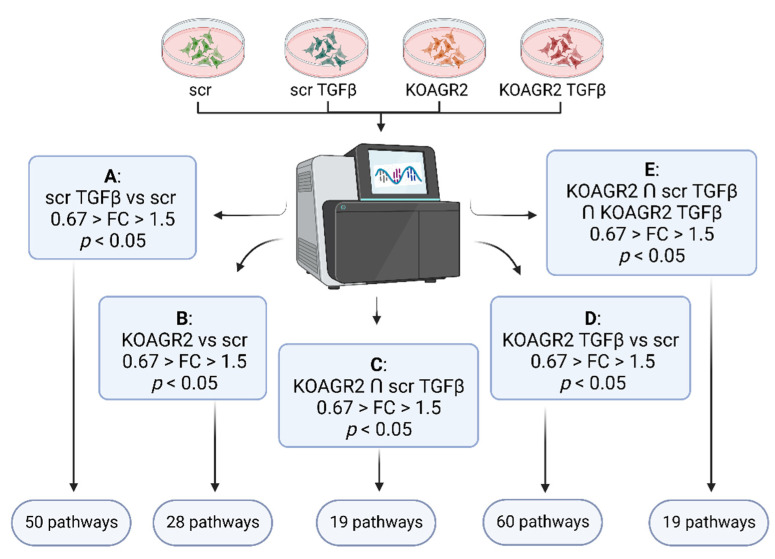
Figure 2.
Schematic representation of samples subjected to RNAseq and the data analysis workflow using DAVID Functional Annotation Clustering tool at the DAVID 2021 Knowledgebase. (A) DAVID clustering of transcripts with significant FC in scr TGF-β samples vs scr control samples. (B) DAVID clustering of significant transcripts from KOAGR2 samples vs scr samples. (C) DAVID clustering of significant transcripts common between KOAGR2 and scr TGF-β. (D) DAVID clustering of significant transcripts from KOARG2 TGF-β vs scr. (E) DAVID clustering of significant transcripts common for KOAGR2, scr TGF-β, and KOAGR2 TGF-β. FC, fold change.