Table 3.
Predicted binding pose of structures 4cZ, 4dE, 4hE, 5bE, and 5fE at the colchicine binding site of α and β tubulin (crystal structure from PDB code: 1SA0) *.
| Compound | Binding Pose and Interactions | Type of Interaction | Active Residues |
|---|---|---|---|
| 4cZ |
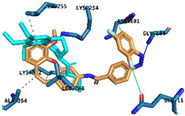
|
Hydrogen Bonds | Gly144 Lys253 |
| Halogen Bond | Glyn11 | ||
| 4dE |
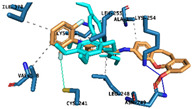
|
Hydrogen Bonds | Asn249 Lys254 |
| Halogen Bond | Cys241 | ||
| 4hE |
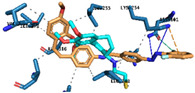
|
Hydrogen Bonds | Asn101 Lys254 |
| π – interaction | Lys254 | ||
| Halogen Bond | Asn101 | ||
| 5bE |
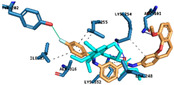
|
Hydrogen Bonds | Asn101 Lys254 Lys352 |
| Halogen Bond | Thr202 | ||
| 5fE |
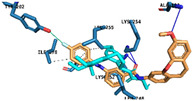
|
Hydrogen Bonds | Ala101 Lys254 Lys352 |
| Halogen Bond | Thr202 |
* As predicted by AutoDock Vina scoring function and active residues interacting with each ligand via halogen bonding, hydrogen bonding o π-interactions. Colchicine extracted from the crystal structure is superimposed and represented as cyan sticks. For clarity, only amino acids that interact with ligands have been shown.
