Abstract
mutS mutators accelerate the bacterial mutation rate 100- to 1,000-fold and relax the barriers that normally restrict homeologous recombination. These mutators thus afford the opportunity for horizontal exchange of DNA between disparate strains. While much is known regarding the mutS phenotype, the evolutionary structure of the mutS+ gene in Escherichia coli remains unclear. The physical proximity of mutS to an adjacent polymorphic region of the chromosome suggests that this gene itself may be subject to horizontal transfer and recombination events. To test this notion, a phylogenetic approach was employed that compared gene phylogeny to strain phylogeny, making it possible to identify E. coli strains in which mutS alleles have recombined. Comparison of mutS phylogeny against predicted E. coli “whole-chromosome” phylogenies (derived from multilocus enzyme electrophoresis and mdh sequences) revealed striking levels of phylogenetic discordance among mutS alleles and their respective strains. We interpret these incongruences as signatures of horizontal exchange among mutS alleles. Examination of additional sites surrounding mutS also revealed incongruous distributions compared to E. coli strain phylogeny. This suggests that other regional sequences are equally subject to horizontal transfer, supporting the hypothesis that the 61.5-min mutS-rpoS region is a recombinational hot spot within the E. coli chromosome. Furthermore, these data are consistent with a mechanism for stabilizing adaptive changes promoted by mutS mutators through rescue of defective mutS alleles with wild-type sequences.
Comparisons of nucleotide sequences encoding enzymes involved in DNA metabolism have revealed remarkable levels of sequence conservation among geographically disparate populations of bacteria. That is, although random mutagenesis of the catalytic site of DNA polymerases has revealed a highly plastic nucleotide-binding domain, very little variation at the sequence level can be documented in the same nucleotide-binding sites among naturally occurring enteric isolates (44). Lack of sequence heterogeneity among these genes is counterintuitive to the diversity that one would expect given a universal mutation rate of 10−9 substitutions per base per generation in the prokaryotic genome (15). As noted by Patel and Loeb (44), 100 million years of eubacterial evolution should have easily allowed mutations to have occurred at every nucleotide position in these loci. They went on to propose that horizontal transfer among bacterial strains could account for the high degree of sequence homogeneity observed in the genomes of feral populations (44).
This hypothesis becomes more intriguing in light of the fact that mutators, cells capable of accelerating bacterial mutation rates 100- to 1,000-fold, constitute more than 1 in 100 isolates of pathogenic Escherichia coli and Salmonella enterica (31). In addition to increasing the basal mutation rate of the genome, mutators defective in methyl-directed mismatch repair (MMR) can also increase the horizontal exchange of DNA, as it is one barrier to recombination between diverged DNA sequences (46). Paradoxically, while MMR− mutators can increase total genomic diversity, they can also homogenize sequences via recombination. Horizontal exchange may afford a mutator cell the opportunity to generate a wild-type sequence, allowing the cell to escape the hypermutable phenotype with a more genetically fit sequence (10, 44). Specifically, mutS-defective MMR mutators, which are the mutators most often found in nature, may play a role in this process by enhancing homeologous recombination following horizontal gene transfer (10).
Horizontal transfer of DNA is acknowledged to be a significant mechanism for the generation of genomic diversity in E. coli (30, 41), although its effect on population dynamics remains a matter of debate (25, 36, 37). In addition to plasmid-borne DNA (which can be frequently exchanged between strains [5]), a significant portion of the E. coli chromosome is thought to have been acquired through horizontal exchange. Conservative estimates place that fraction at between 10 and 16% among natural strains (reviewed in references 9 and 41). Within the chromosome, numerous loci have been identified that have phylogenetic histories decoupled from the organism that contains them. Genes in the rfb complex (responsible for O antigen synthesis), gnd (6-phosphogluconate dehydrogenase, adjacent to rfb), and the hsd genes (under strong destabilizing selection pressure due to their role in type 1 host modification and restriction) are evolutionarily “scrambled,” having undergone repeated, multiple recombination events (1, 3, 34). Likewise, several other loci have been identified that seem to be the result of recombination among natural isolates of E. coli. By examining the patterns of nucleotide polymorphisms as well as the levels of phylogenetic incongruence among gene sequences and large-scale chromosomal measures (e.g., multilocus enzyme electrophoresis [MLEE], randomly amplified polymorphic DNA [RAPD], and restriction fragment length polymorphism [RFLP]) (13, 28, 33), icd (isocitrate dehydrogenase), trpC (N-[phosphoribosyl] anthranilate isomerase-indole-3-glycerol phosphate synthase), pabB (para-aminobenzoate synthase), aceK (isocitrate dehydrogenase kinase), and putP (proline permease) were each shown to have undergone some level of recombination in natural populations of E. coli (33, 39, 53). Moreover, several genes associated with bacterial virulence, e.g., hly (α-hemolysin), kps (type II capsule), sfa (S-type fimbrial adhesin), and pap (P-type fimbrial adhesin), revealed clustered distributions among E. coli reference isolates (4) and exhibited patterns indicative of horizontal transfer. In addition, fliC, encoding the flagellar H antigen, appears to have recombined between O157:H7 and O128:H7, two distantly related serotypes of pathogenic E. coli (47). Most recently, phylogenetic mapping approaches have demonstrated the acquisition of several virulence traits (e.g., intestinal adhesion) in parallel across evolutionarily disparate lineages of enterohemorhagic (EHEC) and enteropathogenic (EPEC) E. coli (48).
We showed previously that the mutS-rpoS region, at 61.5 min on the E. coli chromosome, is another genomic region subject to rearrangement by horizontal exchange. This segment contains what is designated herein an unusual region (UR), consisting of a 2.9-kb stretch of DNA found in E. coli O157:H7, Shigella dysenteriae type 1, and several E. coli reference collection (ECOR) strains (32). The region, in general, is unusual in being punctuated by significant size and sequence polymorphisms between E. coli K-12 and O157:H7 and S. dysenteriae type 1, the last of which contains the insertion sequence IS1 in place of the prpB locus (32). These findings led to the notion that this region may have been forged predominantly by recombination between strains of E. coli and/or S. dysenteriae type 1 (32). A recent study of the mutS-rpoS region in uropathogenic E. coli strains reported a novel sequence of 2.1 kb in the position of the 2.9-kb insert (12). This observation serves as yet another sign of the role of horizontal transfer in the evolution of the region.
In order to test the idea that horizontal exchange is a driving force behind the homogenization of particular sequences on the chromosome of E. coli, we chose to analyze the evolutionary history of the mutS gene in a diverse collection of E. coli strains. The mutS gene is an attractive candidate for this analysis because of its unique role in the formation of mutators with relaxed recombination barriers as well as its physical proximity to a known polymorphic segment of the E. coli chromosome (32). A phylogenetic approach was adopted that compared gene phylogeny to strain phylogeny, making it possible to identify strains in which mutS or any adjacent sequences have recombined. Additionally, any observed phylogenetic discordance between sequences was subjected to rigorous statistical analysis using the incongruence length difference (ILD) test (19).
Our phylogenetic analysis of the mutS gene was aided in two ways. First, the ECOR collection, divided into five phylogenetic groups (A, B1, B2, D, and E), allowed us to capture a broad range of genetic diversity in E. coli (42). Second, partial mutS nucleotide sequences of some ECOR strains were already present in GenBank and were found to contain the conserved ATP-binding domain. We designed primers spanning this region and were able to amplify homologous sequences. The combination of these two factors allowed us to analyze over 100 naturally occurring and pathogenic strains in the present study. This investigation has allowed (i) an evaluation of the extent to which the mutS gene has been horizontally transferred within and between natural and pathogenic classes of E. coli and S. dysenteriae type 1, (ii) the most parsimonious description of the acquisition of several genomic features unique to the mutS-rpoS region of the chromosome among naturally occurring E. coli isolates, and (iii) the identification of a potential cross-over event between the mutS gene and sequences located in the adjacent UR. The implication that mutators may be responsible for the promiscuity of a gene that plays a critical role in their phenotypic etiology is discussed.
MATERIALS AND METHODS
Bacterial strains.
A total of 74 bacterial strains encompassing 66 isolates of E. coli and eight isolates of S. dysenteriae type 1 were included as sources of DNA. Forty-three of the E. coli strains originated from the ECOR collection, which is recognized as representing the extent of genetic variability of the species (42). The remaining 23 strains represent various serotypes and pathogenic classes of diarrheagenic E. coli that have originated from clinical and contaminated food sources (23). EHEC isolates included seven strains of serotype O157:H7 (FDA484, FDA486, FDA488, 95-001, 93-111, 86-24, and FRIK583) and one O26:H11 strain (FDA400). The remaining disease classes of pathogenic E. coli were represented by the following strains: enterotoxigenic E. coli (ETEC) O148:H28 (FDA319), O25:K98 (FDA329), O78:H11 (FDA320 and ATCC35401), and O111 (ATCC43887); enteroinvasive E. coli (EIEC) O152 (FDA162), O143 (FDA164), O136 (FDA269), and O124:NM (ATCC43893); and EPEC O55:NM (FDA321), O127 (FDA322), and four O55:H7 strains (DEC5B through DEC5E). S. dysenteriae type 1 was represented in the study by eight strains (FDA377, FDA567, ATCC20130, ATCC20132, ATCC20011, ATCC20020, ATCC20133, and ATCC20174).
Preparation of bacterial DNA.
Genomic DNA was isolated from bacterial strains using a commercially available extraction matrix (Bio-Rad). Briefly, cells were washed in saline, resuspended in Instagene DNA purification resin, and incubated at 56°C for 30 min. Cell preparations were then vortexed vigorously, incubated at 100°C for 10 min, and centrifuged at 14,000 rpm for 6 min. The remaining supernatant, containing total genomic DNA, was decanted into a sterile microtube.
PCR amplification and sequencing.
PCRs were prepared by adding 20 μl of DNA template, 10× PCR buffer containing 1.5 mM MgCl2 (Perkin-Elmer), 2.5 mM deoxynucleoside triphosphate mix (Pharmacia), and 1.5 U of Taq DNA polymerase (Promega). Oligonucleotide primer pairs used to amplify a segment of the mutS gene and a segment of the mutS-proximal UR in E. coli were added to a final amount of 50 pmol and included mutS primers msec1F (5′-TGCTGAACGAGCCATTTATCGC-3′) and msec1R (5′-TTGGCGACGCCTTCCATTTTCT-3′) and UR primers prpur1F (5′-TTGATACCGGATCGCCGAAAA-3′) and prpur2R (5′-GAACAAGATGATTTGTCCACGT-3′). Amplification of mutS sequences was performed in a PTC-200 thermal cycler (MJ Research) under the following conditions: initial denaturation at 94°C for 5 min; 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min; and final incubation at 72°C for 10 min. The segment of the mutS gene amplified corresponded to base pair coordinates 1859 to 2318 of the mutS coding region in E. coli O157:H7 (GenBank accession no. U69873) and included the conserved ATP-binding domain, which lies in the COOH-terminal half of the MutS protein. Amplification conditions for the UR sequence were an initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 53°C for 1 min, and 72°C for 1.5 min, and ending with incubation at 72°C for 10 min. The UR segment amplified corresponded to base pair coordinates 783 to 1069 of the entire prpB UR sequence of the prpB UR in E. coli O157:H7 (accession no. AF054420). Products from PCR amplification of bacterial DNA were purified and concentrated using Qiaquick spin columns (Qiagen). Nucleotide cycle sequencing was performed in both directions directly on purified PCR templates by the dideoxy chain termination method using Thermo-Sequenase (United State Biochemicals) and the primers described above. Sequence data were then analyzed and assembled using the Genetics Computer Group (University of Wisconsin–Madison) sequence-handling program (14).
Sequence alignment and phylogenetic analysis.
In addition to the 74 mutS alleles sequenced here, another 30 partial mutS sequences, originating from 29 ECOR strains and a single strain of O157:H7 (FDA536, previously called EC536 [31]), were acquired from GenBank and also included in the analysis. ECOR mutS sequences were submitted by E. Denamur, Hospital Robert Debré, Paris, France, under accession numbers AF001987 through AF002010, AJ005826 through AJ005828, AF004287, and AJ242620. The single E. coli O157:H7 sequence (accession no. U69873) was generated previously in our laboratory. All nucleotide sequences were aligned using Clustal X (52). Transitions and transversions in the alignment matrix were assigned equal character weights. Genetic distances between all nucleotide sequences were calculated using the method of Jukes and Cantor (29).
The nucleotide matrices were subjected to phylogenetic analysis by using a total of 380 and 257 bp for mutS and UR, respectively. The phylogenetic method employed uses the principle of maximum parsimony (17) and is available in the program NONA v.2.0 (24). This program was chosen largely for its speed in seeking out the most parsimonious trees, as other parsimony programs became computationally inefficient with the number of sequences analyzed here. Winclada v.0.9.9b (40) is a Windows interface for parsimony analysis and was used to visualize and further analyze resultant phylogenies. The most parsimonious trees were sought using heuristic search methods combined with tree bisection-reconnection branch swapping and random-addition order of sequences. When necessary, a strict consensus method was applied in order to reduce the number of equally parsimonious cladograms into a single tree so that every relationship present in the consensus tree was found in each of the original trees (21). Tree branches with a length of zero were collapsed into polytomies. In these cases, a conservative approach was adopted in which strain polytomies originating from the same node were grouped together as a single clade. All mutS cladograms were rooted using an outgroup sequence derived from Salmonella enterica serovar Typhimurium SL1344 (accession no. M18965) (26). Character support for internal tree nodes was determined by 5,000 iterations of bootstrapping (20) and is available in Winclada v.0.9.9b (40). Relative levels of homoplasy were measured among trees using two separate tree indices, the consistency index (CI) (21) and the retention index (RI) (18). ILD testing (19) was used to measure statistical significance in observed discordance between mutS/mdh and mutS/UR phylogenies. The ILD test evaluates the null hypothesis of congruence between phylogenetic data sets (e.g., nucleotide sequence alignments) (19). Congruence (phylogenetic concordance) is tested by combining two data sets (Ax and Bx) into a single matrix (ABx) from which sample submatrices (Ay and By), identical in size to the two original matrices, are created and partitioned. If the sum of the lengths of subsequent parsimony trees Ay and By is significantly longer (P < 0.05) than the summed length of the original trees (Ax and Bx), more discordance is present between the two sets of data than can be explained by chance alone, and the hypothesis of congruence is rejected (19, 33). ILD tests were performed with 1,000 data partitions using simple heuristic searches. Two other sequence data sets were used as evolutionary standards in the ILD test. The gnd locus, scrambled by multiple recombinations, was used as a known marker for incongruence, while gapA, reiterative of MLEE ECOR phylogeny (38), was used as a known marker for congruence. The version of the ILD test employed here is available in PAUP (Phylogenetic analysis using parsimony) v.4.03b (51).
Colony hybridization.
Four oligonucleotide-length probes were designed and used to investigate the distributions and structural similarities of the mutS-rpoS region. Two of these probes, MRJR and BL129, were chimeric in design and used to identify junction sequences between the prpB locus and the E. coli O157:H7-specific UR (MRJR) as well as UR and rpoS (BL129). Two additional probes, BL148 and F23, complement sequences internal to UR (BL148) and a region upstream of the mutS gene but downstream of fhlA (F23). The sequences of these probes were: MRJR (5′-CGGCCTCATTACTTTATTTTAT-3′), BL129 (5′-GGCCTTTTTCTTTTGTTTGGG-3′), BL148 (5′-GACATATTCGGCAACTGCAC-3′), and F23 (5′-AGATGTGGTTATACTCGATCA-3′). The filter preparations and hybridization conditions were as previously described by Cebula and Koch (8), as modified by Cebula (7).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been deposited in the GenBank sequence database with accession numbers AF291185 through AF291258 for mutS sequences and AF291259 through AF291264 for UR sequences.
RESULTS
Phylogenetic analysis of the E. coli mutS gene.
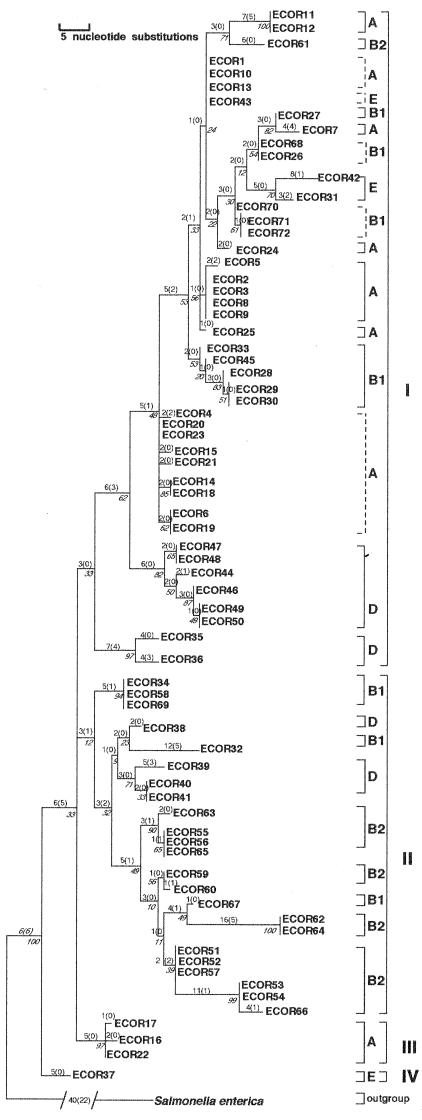
Phylogenetic analysis of a 380-bp segment of the mutS gene from 72 ECOR strains yielded eight equally parsimonious trees, which are summarized as a single strict consensus tree (Fig. 1). This consensus tree generated two important findings. First, four distinct phylogenetic lineages or clades of E. coli strains, designated clades I to IV, were resolved and appear to have diverged from at least two deep radiations in the tree. Second, the five separate lineages of ECOR strains (A, B1, B2, D, and E) appear to be evolutionarily disjunct (polyphyletic) when viewed in the context of mutS phylogeny. These five groups were originally distinguished in a whole-chromosome MLEE neighbor-joining phylogeny (28, 53) and reiterated in a combined MLEE-RAPD-RFLP nonnucleotide parsimony tree (33). In every case, ECOR strains from similar whole-chromosome groups are dispersed into disparate clades on the mutS tree: group A is broken into eight subgroups distributed among clades I and III, B1 into seven subgroups distributed among clades I and II, B2 into five subgroups distributed among clades I and II, D into four subgroups distributed among clades I and II, and E into three subgroups distributed among clades I and IV. Taken together, these findings indicate striking levels of topologic discordance between the phylogenies of mutS and the E. coli chromosome.
FIG. 1.
Phylogenetic relationships of mutS alleles from 72 ECOR strains. The tree shown represents the strict consensus of eight equally parsimonious trees and has a tree length of 283 steps. Most parsimonious trees had a CI of 0.48 and an RI of 0.83. Measures of clade confidence are reported in italics below each node in the form of bootstrap values. The internal brackets to the right of each clade reflect monophyletic strain groupings with respect to the E. coli MLEE lineages A, B1, B2, D, and E. Broken internal brackets indicate groups of strains that formed polytomies on the tree and that are ambiguous with respect to strain polyphyly. The larger external brackets designate the four distinct clades derived from mutS sequences (denoted by roman numerals I to IV). Individual branch lengths are presented above each branch; the numbers of unambiguous substitutions that mapped to the tree only once are given in parentheses.
Presumably, these phylogenetic differences are the result of numerous horizontal genetic exchanges of mutS alleles that have accumulated throughout the evolution of the species. It should be noted that, while bootstrapping was instrumental in supporting more terminal relationships in the mutS tree, it did not support structure among all deep nodes with equal levels of confidence. This observation, however, is simply a result of the limited number of nucleotide substitutions that gave rise to these specific clades but were lost during subsequent bootstrap iterations. Despite this nuance, the most parsimonious solution of these data remains one that supports a mutS phylogeny highly discordant with ECOR strain phylogeny.
Phylogenetic discordance between mutS and mdh.
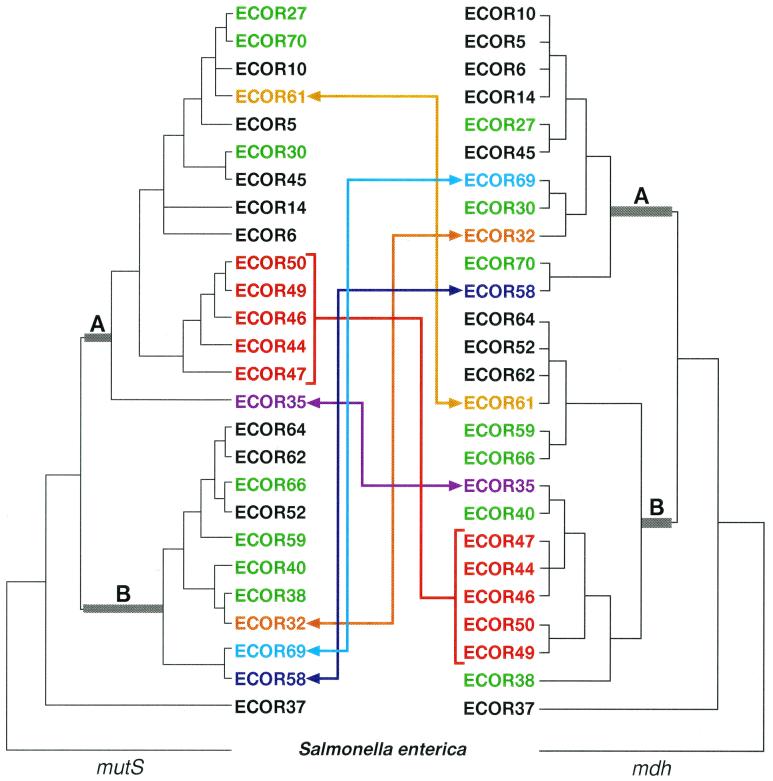
In order to investigate further the extent to which mutS may be phylogenetically decoupled from the strains that possess them, the mutS tree was compared to a known whole-chromosome anchor locus. Malate dehydrogenase (mdh) is one of several housekeeping genes that appear to be clonally inherited along with the E. coli chromosome and, as a result, yields a phylogeny largely concordant with the ECOR whole-chromosome tree (6, 33, 45). A one-to-one phylogenetic comparison of 27 strains for which an mdh phylogeny has previously been reported (45) demonstrated a substantial number of ECOR strain incongruences between the two genes (Fig. 2). Visual inspection of the two topologies revealed 10 ECOR strains located in two entirely separate lineages (A and B) among the two trees, with 5 of these 10 strains (ECOR 44, 46, 47, 49, and 50) appearing to have recombined en masse, possibly due to a single horizontal transfer event. In addition to these major interclade transpositions, a number of subtler intraclade differences were also observed with ECOR strains 27, 70, 30, 66, 59, 40, and 38, all showing topologic discrepancies between the two loci (green strains, Fig. 2). While this latter group of strains did not exhibit major translocations between the two trees, the possibility that the mutS gene has recombined in these strains cannot be excluded. Since closely related strains have highly homologous sequences, the horizontal exchange of DNA between them will have little effect on tree topology and consequently may go unnoticed.
FIG. 2.
Phylogenetic comparisons of mutS and mdh among ECOR strains. The colored arrows mark the lateral movement of strains between distinct mutS and mdh clades (designated A and B). The two clades in which strains have been displaced are marked with thickened, gray-shaded basal branches. Strains depicted in green represent within-clade differences in the topology of the two trees. The red bracket denotes a group of strains that potentially acquired the same mutS allele. The gold, purple, navy, light blue, and orange arrows each connect an ECOR strain that has demonstrated interclade movement relative to the mutS and mdh phylogenies. The mutS tree was derived from the most parsimonious tree presented in Fig. 1, and the mdh tree was derived from a previously reported phylogeny (45). Both trees were rooted with S. enterica serovar Typhimurium as the outgroup.
These observations were further supported by ILD testing, which tests the null hypothesis of congruence between two genes (19). The two genes demonstrated significant incongruence (P < 0.001 for 1,000 partitions) when all 27 strains were included in the test. Only after the removal of 10 strains did the test fail to yield significant ILD values between the two genes (n = 17, P < 0.10 for 1,000 partitions). In other words, the removal of ECOR strains 35, 44, 46, 47, 49, 50, 59, 61, 69, and 70 was necessary to derive a mutS phylogenetic tree that was statistically congruent with the mdh tree. Surprisingly, it was not necessary to remove ECOR strains 32 and 58 to achieve statistical congruence between the two phylogenies. ILD sensitivity to incongruous strain relationships was confirmed using sequences from two additional genes, gnd and gapA (33, 38). As expected, gnd-mdh comparisons yielded significantly discordant P values (n = 11, P < 0.001), while gapA-mdh comparisons revealed an expected concordance among phylogenetic signals (n = 9, P < 1.00). In this comparative analysis, both visual inspection of the mutS and mdh phylogenies and statistical testing employing an ILD approach supported the hypothesis that the E. coli mutS gene has a phylogenetic history that is decoupled from the evolution of the strains in which it resides.
Phylogenetic relationships of mutS alleles from pathogenic E. coli and S. dysenteriae type 1.
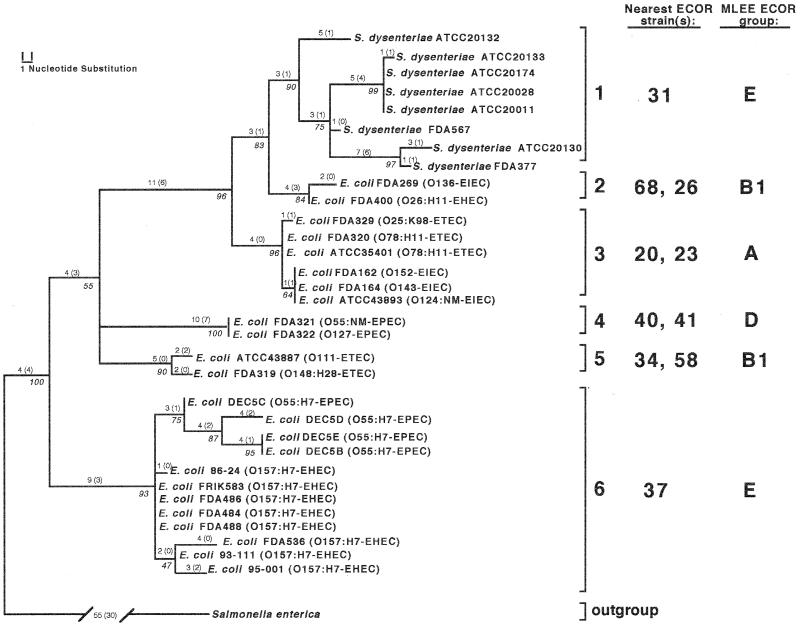
The mutS sequences from 32 pathogenic E. coli strains representing four distinct pathogenic classes were combined with sequences from eight strains of S. dysenteriae type 1 and subjected to the maximum parsimony methods described above. Three equally parsimonious trees resulted from this analysis and were collapsed into the consensus tree shown in Fig. 3. This combined-pathogen analysis yielded several interesting observations. First, pathogenic strains sorted into six distinct clades (denoted 1 through 6), with each of these groups arising from a different location in the mutS ECOR tree presented in Fig. 1. This observation is reinforced by the fact that each of the six pathogenic clades retained a different ECOR strain(s) as its nearest phylogenetic neighbor. Second, with the exception of S. dysenteriae type 1, which solely constitutes clade 1, none of the four pathogenic classes of E. coli demonstrated strain monophyly. Surprisingly, three of the six clades comprised strains from at least two different pathogenic E. coli classes—clade 2 is composed of EIEC and EHEC strains, clade 3 contains both EIEC and ETEC strains, and clade 6 is composed of EHEC and EPEC strains. Third, several groupings within the mutS tree tend to support previous findings with regard to the evolution of at least two of the pathogenic E. coli lineages examined here. For example, all of the O157:H7 strains cluster closely together with O55:H7 strains, and both have ECOR37 (an E group strain) as their nearest neighbor. This is consistent with the MLEE-based hypothesis that a strain from the classic serotype O55:H7 may have given rise to the modern O157:H7 lineage of hemorrhagic E. coli, both of which are closely related to ECOR37 (45, 48, 54). Furthermore, the EIEC strains in this study were found to be distributed among ECOR strains representing MLEE groups A and B1. This finding is reinforced by recent ribotype studies on EIEC isolates, which also suggest that enteroinvasive E. coli originated from diverse A, B1 and B2 ECOR lineages (49). Finally, S. dysenteriae type 1 has ECOR31 as its nearest phylogenetic neighbor. Previous taxonomic observations have placed the emergence of S. dysenteriae type 1 from within the ECOR group B1 reservoir (49). While ECOR31 is a group E strain, it should be noted that the mutS allele of ECOR31 also appears to be group B1 in origin (Fig. 1). This would suggest that ECOR31 underwent a punctual recombination with a group B1 strain, acquiring a B1-type mutS allele which it later passed to S. dysenteriae type 1 during emergence of this pathogen from an ancestral E. coli lineage.
FIG. 3.
Phylogenetic relationships of mutS nucleotide sequences from pathogenic E. coli and S. dysenteriae type 1. The tree shown represents the strict consensus of three equally parsimonious trees and has a tree length of 171 steps (CI = 0.71, RI = 0.89). Measures of clade confidence are reported in italics below each node in the form of bootstrap values. Individual branch lengths are presented above each branch, and the numbers of unambiguous substitutions that mapped to the tree only once are given in parentheses. The brackets to the right of the tree indicate the six distinct clades of pathogenic strains (1 to 6). The nearest ECOR strain(s) based on mutS relationships and the MLEE ECOR group designation is listed to the right of each bracketed clade.
Evaluation of the aligned mutS nucleotide sequences from pathogenic E. coli and S. dysenteriae type 1 strains revealed the existence of synapomorphic (shared and derived) nucleotides that are unique to specific pathogenic mutS clades. These signature synapomorphic substitutions, their positions in the Clustal X alignment, and the groups that they distinguish are listed in Table 1. Synapomorphic nucleotides at positions 47, 95, 167, 272, 323, 341, 350, and 353 are unique to EPEC clade 4, which comprised FDA321 and FDA322 of serotypes O55:NM and O127, respectively. In addition, EHEC-EPEC clade 6, composed solely of O157:H7 and O55:H7 strains, share unique positions 234, 236, 264, and 284, while substitutions at positions 17, 218, and 296 distinguished clade 2, which contains a single EHEC (O26:H11) and a single EIEC (O136) strain. Finally, two other clades (1 and 5) are each represented by a single synapomorphic position, 311 for clade 1 and 197 for clade 5. Overall, 17 synapomorphic sites within the specified sequence are unique to a single pathogenic clade of mutS alleles. These substitutions should prove useful in identifying novel isolates of pathogenic E. coli using PCR amplification and colony hybridization techniques.
TABLE 1.
Signature nucleotide substitutions unique to individual mutS clades of pathogenic E. coli strains a
| Clade no. | Nucleotide at position:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17 | 47 | 95 | 167 | 197 | 218 | 234 | 236 | 264 | 272 | 284 | 296 | 311 | 323 | 341 | 350 | 353 | |
| 1 | C/G | C | C | C | A | C | T | A | T | T | T | T | [A] | G | G | T | G |
| 2 | [T] | C | C | C | A | [T] | T | A | T | T | T | [C] | G | G | G | T | G |
| 3 | C | C | C | C | A | C | T | A | T | T | T | T | G | G | G | T | G |
| 4 | C | [A] | [G] | [T] | A | C | T | A | T | [C] | T | T | G | [A] | [A] | [C] | [A] |
| 5 | C | C | C | C | [G] | C | T | A | T | T | T | T | G | G | G | T | G |
| 6 | C/G | C | C | C/G | A | C | [C]b | [G] | [C]b | T | [C] | T | G | G | G | T | G |
Signature nucleotides unique to every member of each clade are bracketed. Nucleotide substitution positions were derived using the Clustal X multiple sequence alignment program (52) and visualized in MacClade v. 3.04 (35). Clade numbers correspond to clades reported in Fig. 3. All other substitutions shown are located in the third position of the codon and are also synonymous.
Synonymous first-position substitution.
Distribution and phylogenetic mapping of other mutS-rpoS region features.
The mutS-rpoS region contains numerous polymorphisms, including the UR sequence, a 2,930-bp stretch of DNA found in E. coli O157:H7 and S. dysenteriae type 1 but absent in K-12 (32). In order to determine structural similarities of the mutS-rpoS region (including UR sequence) across a diverse group of naturally occurring E. coli, colony hybridization experiments were performed on the entire ECOR collection using several oligonucleotide probes. Probe BL148 complements internal UR sequence downstream from mutS present in E. coli O157:H7 and S. dysenteriae type 1 but not in E. coli K-12. F23 recognizes a sequence upstream from mutS adjacent to fhlA (formate hydrogen lyase). This sequence is present in S. dysenteriae type 1 but absent in E. coli O157:H7 and K-12. As shown in Fig. 4, gene order in this region is 5′-fhlA-mutS-prpB-UR-rpoS-3′ on the chromosome of E. coli O157:H7 (32). By employing chimeric probes at the junctions of prpB-UR (MRJR) and UR-rpoS (BL129), signifying UR- and rpoS-proximal sequence, we were able to examine which sequences lie adjacent to one another as well as to inspect the overall distribution of theses features among a diverse collection of E. coli strains. The hybridization analysis revealed that these sequences are not represented across the entire ECOR collection (Fig. 4). Rather, UR sequence (as revealed by probe BL148) appears to be present in only 39% (n = 28) of all ECOR strains, while upstream F23 sequence is present in less than half of all ECOR strains (n = 33, 46%). In addition, an intact prpB-UR junction was found in only five ECOR strains (see probe MRJR in Fig. 4), and the UR-rpoS junction was intact in seven ECOR strains (see probe BL129 in Fig. 4). These data suggested that while UR sequence is present in a large portion of ECOR strains, its positional homology is quite diverse among strains. Observed structural diversity of this nature further supports the notion that the mutS-rpoS region has endured genetic exchange and rearrangement of DNA during the evolution of this portion of the E. coli chromosome.
FIG. 4.
Distribution of four mutS-rpoS region sequences among ECOR strains. The schematic shows the genetic organization of the E. coli chromosome from 61 to 62 min. The bold arrows show the length and direction of the coding regions of the genes indicated. The UR of E. coli O157:H7 and corresponding region in E. coli K-12 are shown, with their sizes given in parentheses. The four probes employed in colony hybridization studies are indicated in ovals and are positioned to show their relative locations on the chromosome. The presence (+) or absence (−) of a sequences is given below each probe for 72 ECOR strains, E. coli O157:H7 and K-12, and S. dysenteriae type 1. The asterisk marks a weakly positive reaction. The map is scaled in 2,000-bp increments based on sequence coordinates of the E. coli K-12 sequence contig Ecu29579, available in GenBank.
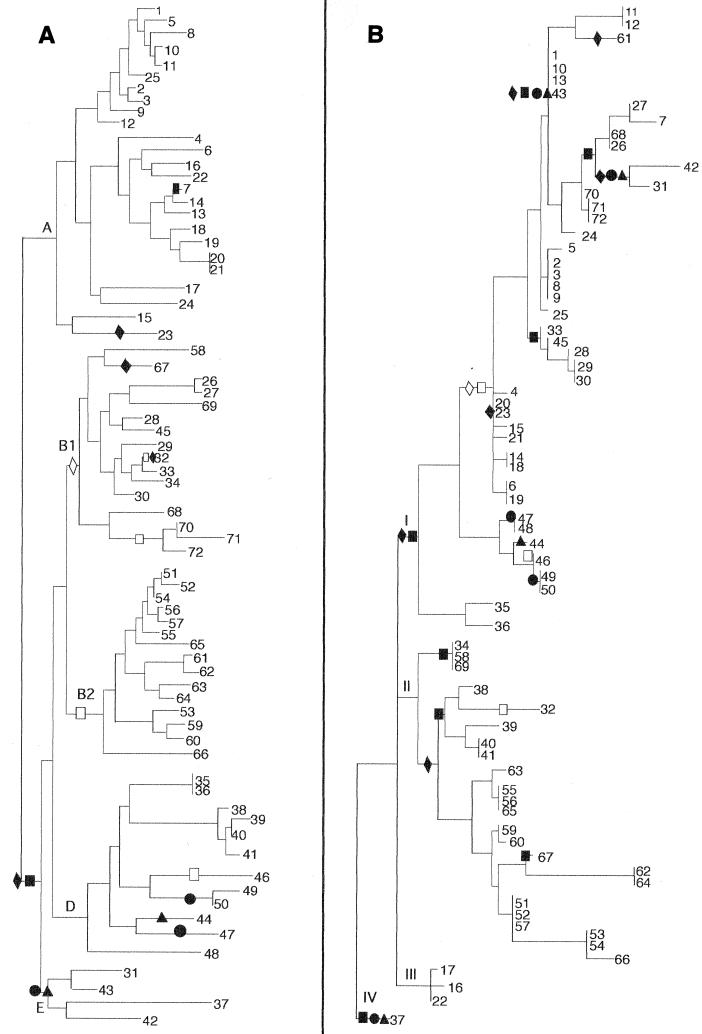
An examination of the phylogenetic history of the probe data in Fig. 4 further supports the notion that the mutS-rpoS region has been subject to horizontal transfer in E. coli. The binary data from the probing analysis were converted into phylogenetic characters and mapped to the MLEE and muts ECOR trees using the principles of maximum parsimony. Various optimization attempts were made in order to determine the least number of times that each of the sequence traits could have arisen in evolution. This approach allowed a description of the most parsimonious distributions for each of the four probes (Fig. 5). Surprisingly, none of the four probe distributions could be accounted for by a single evolutionary event when mapped to the MLEE ECOR tree (Fig. 5A). The minimum number of evolutionary transformations (steps) was observed for probe MRJR (prpB-UR junction), which mapped to the MLEE tree only twice. Conversely, the distribution of UR within E. coli as detected by probe BL148 (internal UR) could be accounted for by no fewer than six steps. Overall, the most parsimonious solution for the four probes required a total of 16 steps. These data suggest that none of the four sequences analyzed here is completely linked to the evolution of any of the others or to the chromosome in general, each showing some level of gain or loss of sequences in parallel across ECOR strain phylogeny. When the same data were mapped to the mutS ECOR tree (Fig. 5B), the most parsimonious scenarios for MRJR and BL148 were increased to 4 and 11 steps, respectively. Combined, the four probes required a total of 27 steps to be optimally mapped to the mutS tree. This increase in steps from the MLEE tree to the mutS tree is not entirely unexpected, since mutS appears to have undergone numerous horizontal exchanges itself. Thus, a least-assumptive scenario of 27 steps is likely a result of numerous polymorphisms that have accrued not only in the evolution of these four probe-specific sequences but also in the unique evolution of the mutS locus.
FIG. 5.
Phylogenetic distributions of four mutS-rpoS region sequences mapped onto E. coli MLEE and mutS phylogenies. Hybridization data were optimized on the (A) MLEE and (B) mutS trees according to the principle of maximum parsimony. The optimization scheme shown represents the most parsimonious scenario for the evolution of these four sequences. In clades for which equally parsimonious optimizations exist, the accelerated transformation scheme is presented (21). Probe sequences are represented on the trees by the following symbols: ▴, MRJR; ●, BL129; ■, BL148; and ⧫, F23. The presence of a symbol represents a single evolutionary transformation (step). Shaded symbols represent the gain of specific sequences along the indicated lineage, while open symbols mark the loss of the sequence from a clade.
Phylogenetic evidence for a recombinational crossover between mutS and the downstream UR sequence.
The tree-mapping data revealed multiple sites for the origination of sequences detected by probe MRJR on the mutS tree (Fig. 5B, triangles), indicating that the mutS-proximal end of the UR may be phylogenetically disjoined from mutS. Since the two sequences are separated by less than 1 kb and this disjunction could result from a recombination event that occurred between the two regions, a phylogenetic analysis was undertaken for the five ECOR strains and two pathogenic strains that possessed mutS and intact prpB-UR junction sequences.
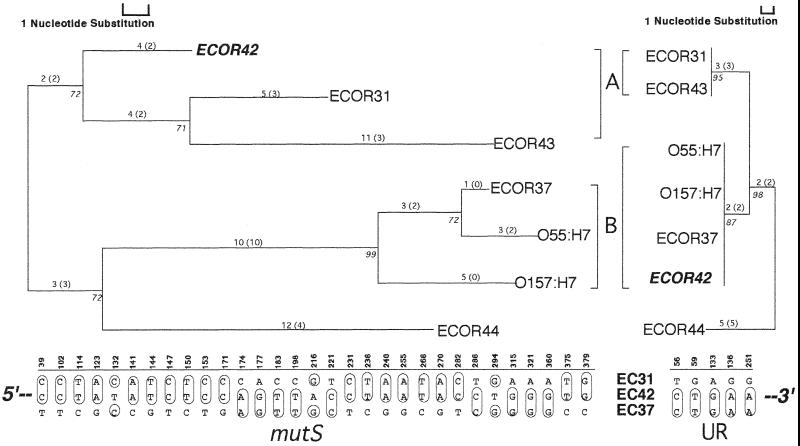
Visual inspection of the resultant mutS and UR trees revealed one key aspect (Fig. 6). ECOR42 appeared in a clade (A) along with ECOR31 and ECOR43 in the mutS tree, but emerged in a disparate clade (B) adjacent to ECOR37, O157:H7, and O55:H7 in the UR tree. This finding is buttressed by examining the genetic similarities between the ECOR42 sequences and the remaining mutS and UR sequences (Table 2). Among mutS sequences, ECOR42 is genetically more similar to ECOR31 in clade A (96.8%) than to clade B member ECOR37 (94.0%). However, in the UR analysis, ECOR42 is 100% identical to clade B members ECOR37, O157:H7, and O55:H7 but 98.1% similar to clade A member ECOR31. Taken together, these data are consistent with the interpretation that a cross-over event has occurred downstream of the mutS sequence but upstream of UR sequence, giving rise to the distinct relationships observed for ECOR42 between the two regions. ILD testing of the two data sets failed to yield a significant difference in congruence for these two phylogenies (P < 0.66 for 1,000 partitions), even though all five polymorphic sites in the UR nucleotide matrix definitively coupled ECOR42 with the other members of clade B (Fig. 6). The failure of the ILD test to respond to topological differences was likely a result of abbreviated tree length (the UR tree was only 12 steps) combined with two groups of mutS substitutions that clustered ECOR42 with ECOR37 and not with ECOR31 (see nucleotides 174, 177, 183, and 198 and 315, 321, and 360 in Fig. 6). Regardless of deficiencies in the statistical rigor of ILD in this instance, phylogenetic analysis identified a single strain (ECOR42) in which the mutS-UR portion of the E. coli chromosome may have recombined.
FIG. 6.
Phylogenetic evidence for a crossover between mutS and the adjacent UR in E. coli. The trees shown represent the most parsimonious phylogenies for mutS and UR among the seven ECOR strains known to possess both mutS and intact prpB-UR junction sequences. The mutS tree shown had a length of 63 steps (CI = 0.76, RI = 0.71). The UR had a length of 12 steps (CI = 1.00, RI = 1.00). The distribution of nucleotide substitutions that gave rise to the topologies is given below each tree for ECOR (EC) strains 31, 42, and 37, and these illustrate the shift in genetic similarities for ECOR42 across the two regions. The numbers above each substitution show the exact position in the Clustal X sequence alignment. The trees shown were midpoint rooted.
TABLE 2.
Genetic similarities among mutS and UR nucleotide sequences between MRJR-positive strains of E. colia
| E. coli strain (no.) | % similarityb to strain no:
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| ECOR43 (1) | 100 | 98.1 | 98.1 | 96.1 | 98.1 | 98.1 | |
| ECOR31 (2) | 96.1 | 98.1 | 98.1 | 96.1 | 98.1 | 98.1 | |
| ECOR42 (3) | 94.6 | 96.8 | 100 | 96.5 | 100 | 100 | |
| ECOR37 (4) | 91.1 | 92.3 | 94.0 | 96.5 | 100 | 100 | |
| ECOR44 (5) | 94.5 | 93.7 | 95.4 | 93.7 | 96.5 | 96.5 | |
| O157:H7c (6) | 93.2 | 92.9 | 93.7 | 98.1 | 93.2 | 100 | |
| O55:H7d (7) | 90.5 | 91.7 | 93.4 | 98.9 | 93.7 | 97.1 | |
Genetic distances were derived from pairwise sequence comparisons using the Jukes-Cantor correction (29).
UR values are given above the diagonal, while mutS values are given below the diagonal.
Represented by strain FDA536.
Represented by strain DEC5C.
DISCUSSION
By combining nucleotide sequence analysis with the techniques of phylogenetic construction, this study demonstrated marked discordance between the phylogeny of mutS alleles and the phylogeny of the E. coli strains in which they reside. Observed differences between gene evolution and strain evolution are readily interpreted as examples of horizontal exchange of the mutS gene between diverse lineages of E. coli (Fig. 1 and 2). While it is possible that mutational convergence in the form of nucleotide reversals and parallelisms (homoplasy) may account for some of the incongruences observed between the mutS and whole-chromosome phylogenies, many of the differences documented among mutS sequences represent radical topologic departures from established ECOR phylogeny. Given the highly conserved nature of this portion of the mutS molecule, these differences could not be accounted for by nucleotide convergence alone. This conclusion is supported by examining the pattern of mutations within mutS that proved to be unique to each of the various clades of pathogenic E. coli. In every case, these substitutions are either silent, third-position substitutions or rare first-position substitutions that remain silent with respect to codon alteration (Table 1).
Previous studies have sought to identify recombinagenic loci in E. coli as being punctual or scrambled, with punctual referring to an isolated recombination event and scrambled denoting the phylogenetic displacement of numerous strains, presumably due to greater levels of recombination (33). In cases of punctual recombination, it is often possible to identify the recipient strain that has recombined from its discordant tree position. In the mutS tree, however, extensive levels of horizontal exchange precluded the identification of many of the recipient ECOR strains. The only exceptions appear to be ECOR67 (MLEE group B1), ECOR32 (MLEE group B1), and ECOR43 (MLEE group E), all of which appear to have recombined individually into other E. coli lineages. In every other case, the repeated lateral transfer of mutS alleles among ECOR strains has made the identification of recipients of recombined mutS alleles an intractable task using tree topology alone. Branch pattern differences among mutS alleles approach levels of incongruence observed in E. coli for genes such as rfb, gnd, and hsd, all of which appear to be evolutionarily scrambled compared to E. coli strain phylogeny (1, 3, 33, 34). In common with these genes, the mutS locus appears to have endured extensive levels of horizontal exchange during its evolution in E. coli. A recent report attempts to delimit the timing and mechanism of evolution of the mutS-rpoS region (27). The authors state “that the genomic region is old,” basing their hypothesis on the similarity of synonymous substitution rates for mutS-rpoS coding sequences compared to other regions of the genome. Their conclusion unfortunately may be confounded by the fact that recombination could have either a diversifying or homogenizing effect on population structure, depending on the relatedness of the strains involved (16). Clearly, closely related E. coli strains would have comparable rates of change at conserved loci, obscuring the detection and timing of such events.
Phylogenetic analysis of the mutS-proximal end of the adjacent UR revealed the existence of a possible crossover event between the mutS gene and UR. Based on the position of ECOR42 in the UR tree, it is likely that the phylogenetic differences observed for this strain are due to the exchange of mutS alleles between isolates. In the UR tree, ECOR42 reunites with other traditional group E members such as ECOR37, O157:H7, and O55:H7 (all members of clade B in the UR tree; Fig. 6). This is in contrast to the mutS analysis, in which the mutS allele from ECOR42 was found to have been replaced with mutS from another strain in MLEE ECOR group B1. Nucleotide sequence analysis of the intervening sequence may yield the precise crossover site between these two regions. At least two aspects of the mutS-rpoS region, however, could make the identification of such sites problematic. First, given the probability of redundant exchanges over homologous stretches of DNA in this region, the detection of a single crossover event seems unlikely. Second, several of these exchanges may have taken place early in the evolution of the species. This could confound the isolation of precise crossover sites, as point mutation tends to ameliorate sites of integration for foreign DNA, although a precise crossover point in this region of the E. coli K-12 and O157:H7 chromosome has recently been localized (32).
The overall significance of a highly recombinagenic mutS to hypermutability and pathogenesis remains to be determined. However, a role for recombination in infection and resultant disease sequelae should be expected. The hypermutable phenotype has recently been implicated in disease progression in the lungs of cystic fibrosis patients (43). Promiscuous exchange via recombination is clearly one outcome of a mutS phenotype and has already been implicated as playing a major role in the evolution of Pseudomonas aeruginosa in environmental and disease habitats (11, 50). Further, recombination has been invoked mechanistically to explain the unusual levels of homogeneity observed in the active sites of key enzymes that may be employed by pathogenic bacteria (44). It is intriguing to speculate on the evolutionary and biological significance of a recombinagenic mutS gene in E. coli populations, particularly in light of the fact that a majority of cells with the hypermutable phenotype are reliant on defective mutS alleles for their maintenance (10). From an evolutionary standpoint, continued survival in a population must favor nonmutators, since there is likely a long-term disadvantage to maintaining a high mutation rate (22). Thus, while mutators seem to become prominent in a population during times of stress, a mechanism would seemingly have to exist to stabilize and restore wild-type MMR function following the generation of individuals more aptly suited for survival in harsh environments. The horizontal exchange of distinct mutS alleles into and out of mutator strains could represent one such mechanism. That is, the rescue of mutS defects may be the driving force for extensive genetic exchange in the region and would account for the striking levels of horizontal transfer observed here. Indeed, recombination provides the most direct route for reversing a mutS mutator phenotype in nature, since most mutS defects thus far described are due to deletions that are not otherwise revertible (31; unpublished results).
In summary, we have presented a phylogenetic description of the mutS gene in E. coli. These data underscore the notion that the mutS-rpoS region, in particular, the mutS gene itself, is a “bastion of polymorphism” (32, 37), possessing an evolutionary history decoupled from that of the chromosome of the organism in which it resides. Evidence of horizontal transfer of mutS alleles denotes yet another segment of the E. coli chromosome that appears to be promiscuous in its origins. It will be interesting to determine the extent to which recombination may have forged the evolutionary histories of other genes in the region, including prpB and rpoS, both of which are involved in the adaptation and survival of the cell. Whatever the final outcome, it is evident that molecular phylogenetic techniques such as those employed here offer valued and predictive insight into the evolution of the chromosome of E. coli.
ACKNOWLEDGMENTS
We are very grateful to M. Allard, A. Benson, M. Kotewicz, and D. Levy for insightful comments; to A. Benson, K. Lampel, H. Ochman, B. Tall, P. Tarr, and T. Whittam for kindly contributing strains; and to K. Nixon and P. Goloboff for making available the Winclada and NONA phylogenetic analysis software packages.
REFERENCES
- 1.Barcus V A, Titheradge A J B, Murray N E. The diversity of alleles at the hsd locus in natural populations of Escherichia coli. Genetics. 1995;140:1187–1197. doi: 10.1093/genetics/140.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, Denamur E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 strains. J Infect Dis. 1998;177:642–50. doi: 10.1086/514217. [DOI] [PubMed] [Google Scholar]
- 3.Bisercic M, Feutrier J Y, Reeves P R. Nucleotide sequence of the gnd genes from nine natural isolates of Escherichia coli: evidence of intragenic recombination as a contributing factor in the evolution of the polymorphic gnd locus. J Bacteriol. 1991;173:3894–3900. doi: 10.1128/jb.173.12.3894-3900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd E F, Hartl D L. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol. 1998;180:1159–1165. doi: 10.1128/jb.180.5.1159-1165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd E F, Hill C W, Rich S M, Hartl D L. Mosaic structure of plasmids from natural populations of Escherichia coli. Genetics. 1996;143:1091–1100. doi: 10.1093/genetics/143.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd E F, Nelson K, Wang F-S, Whittam T S, Selander R K. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA. 1994;91:1280–1284. doi: 10.1073/pnas.91.4.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cebula T A. Allele-specific hybridization and polymerase chain reaction (PCR) in mutation analysis: the Salmonella typhimurium His paradigm. In: Minear R A, Ford A M, Needham L L, Karch N J, editors. Applications of molecular biology in environmental chemistry. Boca Raton, Fla: CRC Press; 1995. pp. 11–33. [Google Scholar]
- 8.Cebula T A, Koch W H. Analysis of spontaneous and psoralen-induced Salmonella typhimurium hisG46 revertants by oligonucleotide colony hybridization: use of psoralens to cross-link probes to target sequences. Mutat Res. 1990;229:79–87. doi: 10.1016/0027-5107(90)90010-2. [DOI] [PubMed] [Google Scholar]
- 9.Cebula T A, LeClerc J E. Hypermutability and homologous recombination: ingredients for rapid evolution. Bull Inst Pasteur. 1997;95:97–106. [Google Scholar]
- 10.Cebula T A, LeClerc J E. DNA repair and mutators: effects on antigenic variation and virulence of bacterial pathogens. In: Brogden K A, et al., editors. Virulence mechanisms of bacterial pathogens. 3rd ed. Washington, D.C.: ASM Press; 2000. pp. 143–159. [Google Scholar]
- 11.Cebula T A, LeClerc J E. Pseudomonas survival strategies in cystic fibrosis. Science. 2000;289:391–392. doi: 10.1126/science.289.5478.391c. [DOI] [PubMed] [Google Scholar]
- 12.Culham D E, Wood J M. An Escherichia coli reference collection group B2-and uropathogen-associated polymorphism in the rpoS-mutS region of the E. coli chromosome. J Bacteriol. 2000;182:6272–6276. doi: 10.1128/jb.182.21.6272-6276.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desjardins P, Picard B, Kaltenbock B, Elion J, Denamur E. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment length polymorphism. J Mol Evol. 1995;41:440–448. doi: 10.1007/BF00160315. [DOI] [PubMed] [Google Scholar]
- 14.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drake J W. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dykhuizen D E, Green L. Recombination in Escherichia coli and the definition of biological species. J Bacteriol. 1991;173:7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farris J S. The logical basis of phylogenetic analysis. In: Platnick N, Funk V, editors. Proceedings of the 2nd Meeting of the Willi Hennig Society: Advances in Cladistics 2. New York, N.Y: Columbia University Press; 1983. pp. 7–36. [Google Scholar]
- 18.Farris J S. The retention index and the rescaled consistency index. Cladistics. 1989;5:417–419. doi: 10.1111/j.1096-0031.1989.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 19.Farris J S, Kallersjo M, Kluge A G, Bult C. Testing significance of incongruence. Cladistics. 1995;10:315–319. [Google Scholar]
- 20.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 21.Forey P L, Humphries C J, Kitching I L, Scotland R W, Siebert D J, Williams D M. Cladistics: a practical course in systematics. Oxford, U.K: Clarendon Press; 1992. [Google Scholar]
- 22.Funchain P, Yeung A, Stewart J L, Lin R, Slupska M M, Miller J H. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics. 2000;154:959–970. doi: 10.1093/genetics/154.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilligan P H. Escherichia coli: EAEC, EHEC, EIEC, ETEC. Clin Lab Med. 1999;19:505–521. [PubMed] [Google Scholar]
- 24.Goloboff P A. NONA v. 2.0 program and documentation. Tucumán, Argentina: INSUE Fundación e Instituto Miguel Lillo; 1997. [Google Scholar]
- 25.Guttman D S, Dykhuizen D E. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 26.Haber L T, Pang P P, Sobell D I, Mankovich J A, Walker G C. Nucleotide sequence mismatch of the Salmonella typhimurium mutS gene required for mismatch repair: homology of mutS and hexA of Streptococcus pneumoniae. J Bacteriol. 1988;170:197–202. doi: 10.1128/jb.170.1.197-202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbelin C J, Chirillo S C, Melnick K A, Whittam T S. Gene conservation and loss in the mutS-rpoS genomic region of pathogenic Escherichia coli. J Bacteriol. 2000;182:5381–5390. doi: 10.1128/jb.182.19.5381-5390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herzer P J, Inouye S, Inouye M, Whittam T S. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism 3. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 30.Lawrence J G, Roth J R. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeClerc J E, Li B, Payne W L, Cebula T A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 32.LeClerc J E, Li B, Payne W L, Cebula T A. Promiscuous origin of a chimeric sequence in the Escherichia coli O157:H7 genome. J Bacteriol. 1999;181:7614–7617. doi: 10.1128/jb.181.24.7614-7617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lecointre G, Rachdi L, Darlu P, Denamur E. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol Biol Evol. 1998;15:1685–1695. doi: 10.1093/oxfordjournals.molbev.a025895. [DOI] [PubMed] [Google Scholar]
- 34.Liu D, Reeves P R. Presence of different O antigen forms in three isolates of one clone of Escherichia coli. Genetics. 1994;138:6–10. [PMC free article] [PubMed] [Google Scholar]
- 35.Maddison W P, Maddison D R. MacClade v. 3.04 program and documentation. Sunderland, Mass: Sinauer Associates; 1994. [Google Scholar]
- 36.Medigue C, Rouxel T, Vigier P, Henaut A, Danchin A. Evidence for horizontal gene transfer in Escherichia coli speciation. J Mol Biol. 1991;222:851–856. doi: 10.1016/0022-2836(91)90575-q. [DOI] [PubMed] [Google Scholar]
- 37.Milkman R. Recombination and population structure in Escherichia coli. Genetics. 1997;146:745–750. doi: 10.1093/genetics/146.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson K, Whittam T S, Selander R K. Nucleotide polymorphism and evolution in the glyceraldehyde-3-phosphate dehydrogenase gene (gapA) in natural populations of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:6667–6671. doi: 10.1073/pnas.88.15.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson K, Selander R K. Evolutionary genetics of the proline permease gene (putP) and the control region of the proline utilization operon in populations of Salmonella and Escherichia coli. J Bacteriol. 1992;174:6886–6895. doi: 10.1128/jb.174.21.6886-6895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nixon K C. Winclada v. 0.9.9b program and documentation. New York, N.Y: Cornell University; 1999. [Google Scholar]
- 41.Ochman H, Lawrence J G, Groisman E A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 42.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1253. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 44.Patel P H, Loeb L A. DNA polymerase active site is highly mutable: evolutionary consequences. Proc Natl Acad Sci USA. 2000;97:5095–5100. doi: 10.1073/pnas.97.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radman M, Matic I, Taddei F. Evolution of evolvability. Ann NY Acad Sci. 1999;870:146–155. doi: 10.1111/j.1749-6632.1999.tb08874.x. [DOI] [PubMed] [Google Scholar]
- 47.Reid S D, Selander R K, Whittam T S. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J Bacteriol. 1999;181:153–160. doi: 10.1128/jb.181.1.153-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reid S D, Herbelin J, Bumbaugh A C, Selander R K, Whittam T S. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 49.Rolland K, Lambert-Zechovsky N, Picard B, Denamur E. Shigella and enteroinvasive Escherichia coli strains are derived from distinct ancestral strains of E. coli. Microbiology. 1998;144:2667–2672. doi: 10.1099/00221287-144-9-2667. [DOI] [PubMed] [Google Scholar]
- 50.Römling U, Schmidt K D, Tümmler B. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J Mol Biol. 1997;271:386–404. doi: 10.1006/jmbi.1997.1186. [DOI] [PubMed] [Google Scholar]
- 51.Swofford D L. Phylogenetic analysis using parsimony (PAUP∗ v.4.03b) program and documentation. Washington, D.C.: The Smithsonian Institution; 1999. [Google Scholar]
- 52.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang F-S, Whittam T S, Selander R K. Evolutionary genetics of the isocitrate dehydrogenase gene (icd) in Escherichia coli and Salmonella enterica. J Bacteriol. 1997;179:6551–6558. doi: 10.1128/jb.179.21.6551-6559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whittam T S, Wolfe M L, Wachsmuth I K, O/rskov F, O/rskov L, Wilson R A. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]