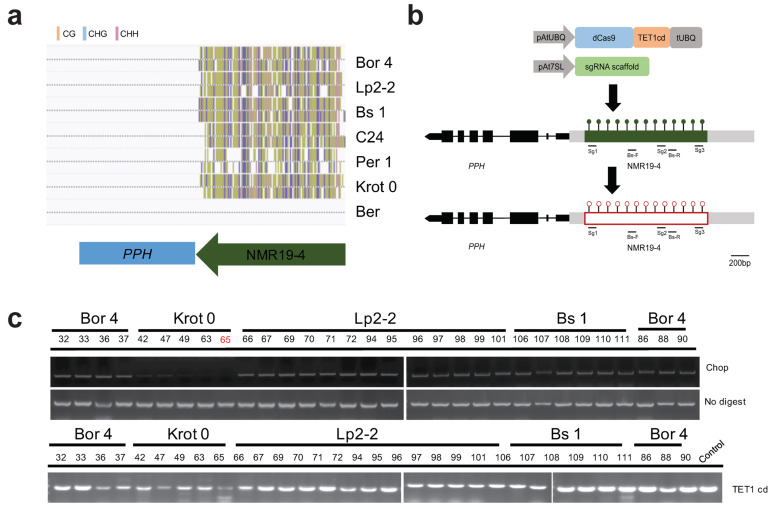
Figure 1.
Targeted DNA demethylation of the Arabidopsis NMR19-4 using the CRISPR/dCas9-TET1cd epigenetic editing system. (a) Methylation level of NMR19-4 in Bor 4, Lp2-2, Bs 1, C24, Per 1, Krot 0, and Ber; (b) Schematic representation of the CRISPR/dCas9-TET1cd system. The target site and the regions for bisulfite PCR (BS-PCR) are shown in the schematic. Green filled circles indicate hypermethylation and red open circles indicate hypomethylation. (c) Chop-PCR analysis of the methylation level of NMR19-4 detected in transformation Arabidopsis T1 generation. The number marked in red represents the line we use in our subsequent experiments.