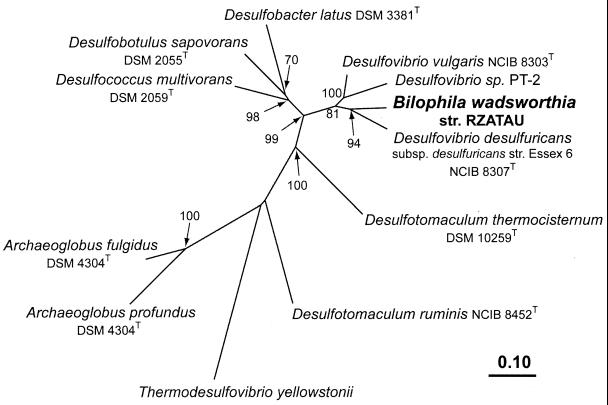
FIG. 3.
Phylogenetic tree reflecting the relationship of the B. wadsworthia RZATAU DSR to the DSRs from Desulfovibrio sp. and other sulfate-reducing microorganisms (21, 45). Tree topology was estimated by using FITCH distance matrix analysis of the concatenated α and β subunit amino acid data sets (546 positions, including D. vulgaris positions 73 to 421 [DsrA] and 18 to 257 [DsrB]). Parsimony analysis was used to determine bootstrap values with an identical data set. Bootstrap values are only indicated for branches, which were recovered in the majority of bootstrap replicates (>50%). The scale bar indicates the number of expected amino acid substitutions per site per unit of branch length. GenBank accession numbers used: U58114, U58115, U58118 to U58127, M95624, AF071499, AF074396, U16723, AF273034, and AF269147.