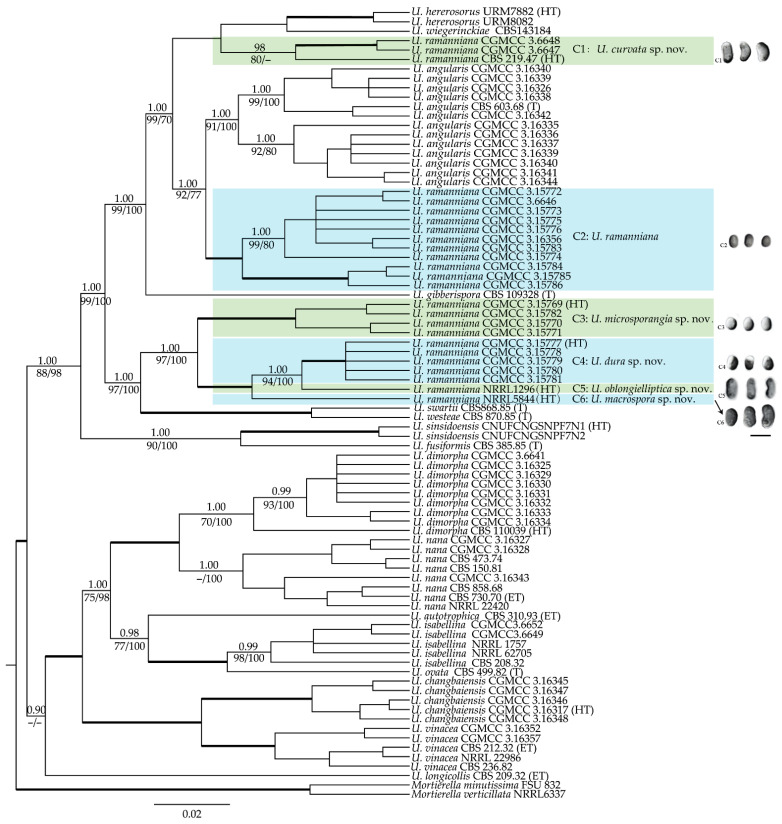
Figure 1.
Phylogenetic tree for Umbelopsis based on a combined data matrix comprised alignments of nSSU, ITS, nLSU, act1, MCM7 and cox1 generated from Bayesian analyses with Mortierella as outgroups. Values above the branches represent significant Bayesian posterior probability values (BPP ≥ 0.95), and values below the branches are maximum likelihood bootstrap proportion (MLBP ≥ 70%) and maximum parsimony bootstrap support values (MPBS ≥ 70%). Branches in bold indicate strong support (MLBP: 100%, MPBS: 100%, BPP: 1.00). Missing or weakly supported nodes (MLBP < 70%, MPBS < 70% or BPP < 0.95) are denoted by a minus sign “−”. The bar at the lower left indicates 0.02 expected changes per site. The new species are highlighted in green and blue. The sporangiospores of novel species established in this study are illustrated on the right side of the tree (scale bar = 5 µm) and correlated with each clade of the U. ramanniana complex using the same clade numbers. U. = Umbelopsis. T = ex-type strain, ET = ex-epitype strain and HT = ex-holotype strain.