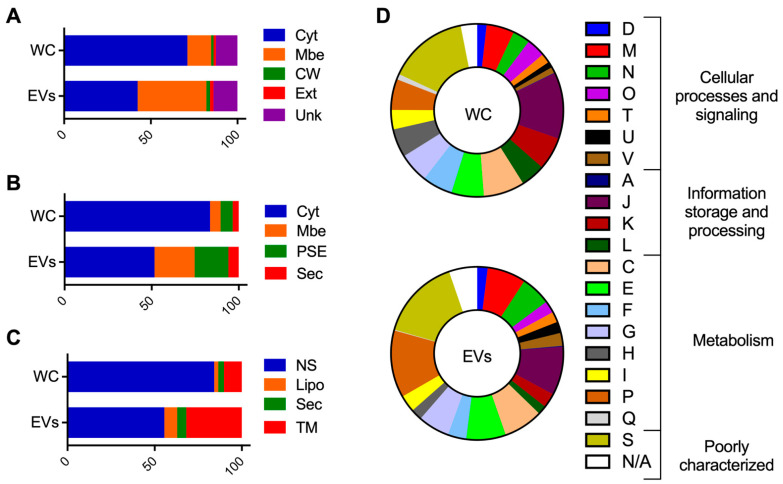
Figure 2.
Comparative proteome analysis of S. aureus HG003 WC and EVs. The prediction of subcellular locations of proteins with PSORTb (A), SurfG+ (B), and PRED-LIPO (C), and the prediction of COG functional categories of proteins with eggNOG-mapper (D). Cyt, cytoplasmatic; Mbe, Membrane; CW, cell wall; Ext, extracellular; Unk, unknown; PSE, surface-exposed; Sec, secreted; NS, no signals found; Lipo, lipoprotein; TM, transmembrane; COG functional categories: D, Cell cycle control, cell division, chromosome partitioning; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, post-translational modification, protein turnover and chaperones; T, signal transduction mechanisms; U, intracellular trafficking, secretion and vesicular transport; V, defense mechanisms; A, RNA processing and modification; J, translation, ribosomal structure, and biogenesis; K, transcription; L, replication, recombination, and repair; C, energy production, and conversion; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; S, function unknown; NA, not available.