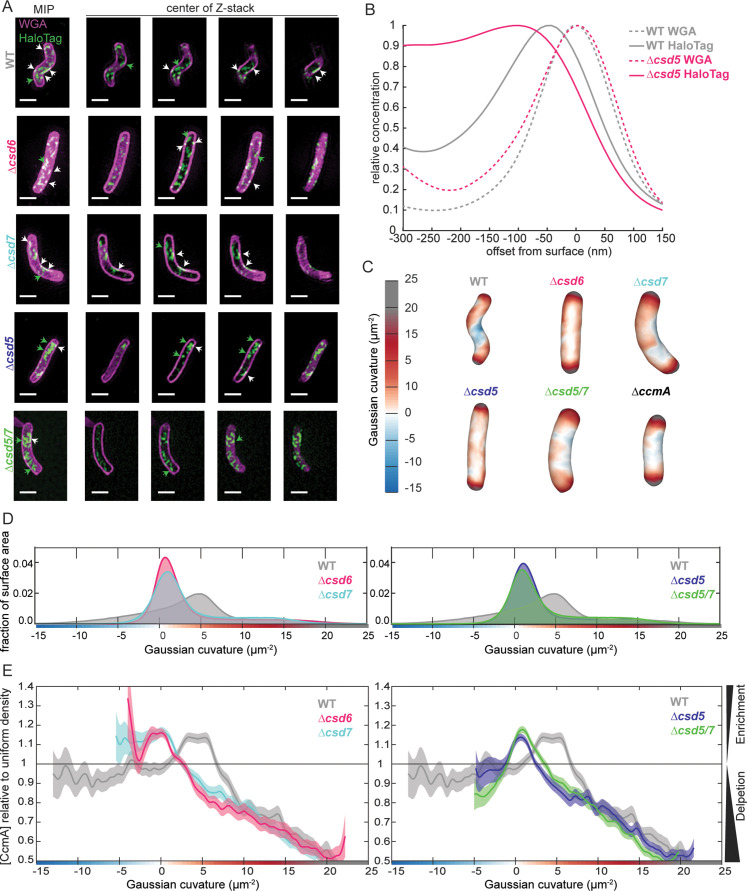
Figure 7. Csd5 is required for CcmA-HaloTag to localize to the cell envelope at the major helical axis.
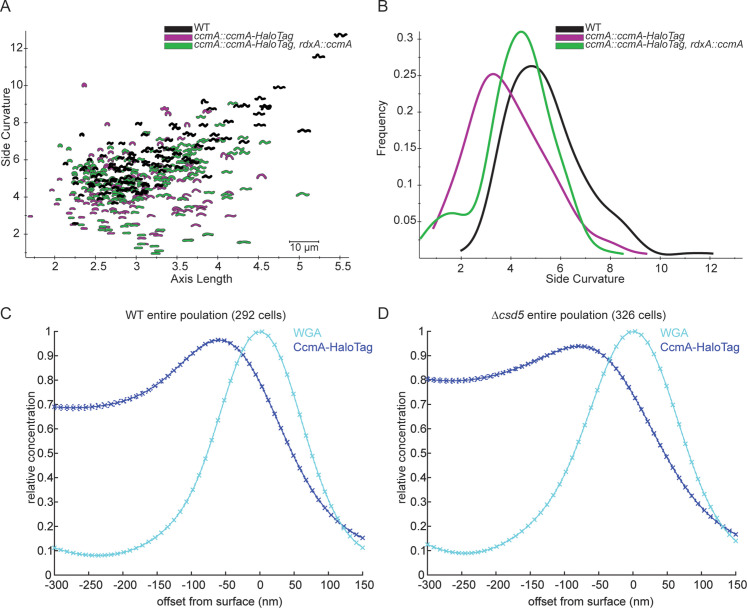
(A) Maximum intensity projections (MIPs) and frames from Z-stacks of H. pylori cells labeled with wheat germ agglutinin (WGA) to label the cell wall (magenta) and JF549 ligand to label CcmA-HaloTag (green), pixels are white where the two signals colocalize. Green arrows indicate cytoplasmic CcmA-HaloTag. White arrows indicate CcmA-HaloTag signal that colocalizes with the cell envelope. Scale bars = 1 µm. (B) Representative plots displaying the relative concentration of CcmA-HaloTag and WGA signal from one WT and one ∆csd5 cell at the cell surface (offset from surface = 0) and at computationally generated cell surfaces that are inside (offset from surface < 0) and outside (offset from surface > 0) of the cell surface. (C) Representative H. pylori cells from each population with Gaussian curvature values mapped on the cell surface. (D, E) Top: histogram of Gaussian curvature of cell surfaces of each population. Bottom: surface Gaussian curvature enrichment of relative concentration of CcmA signal in each population where 1 = uniform concentration of CcmA, shaded regions indicate SEM. In panels (C–F), WT n = 292, ∆csd6 n = 264 cells, ∆csd7 n = 355, ∆csd5 n = 326, ∆csd5/7 n = 523. Strains used: SSH39B, SSH49A, SSH50A, JS09, SSH70A, and LSH117.