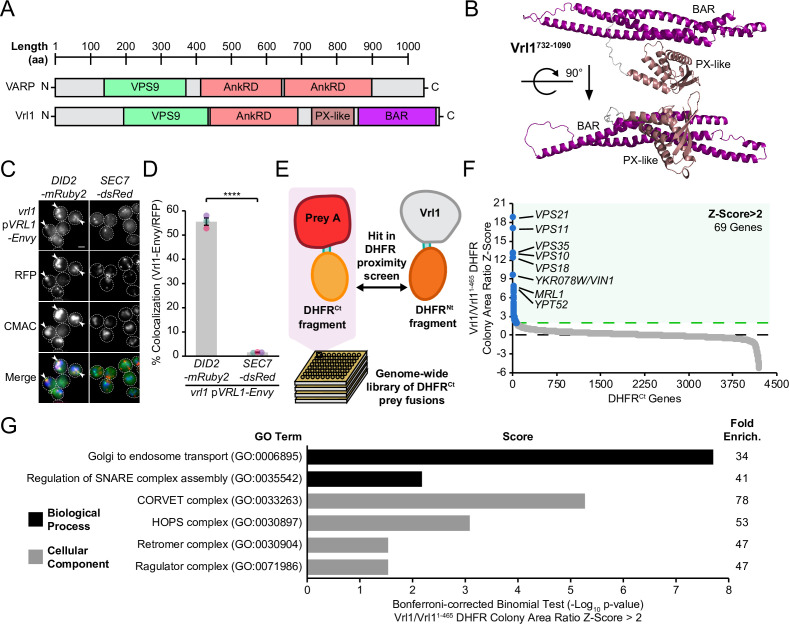
Figure 1. Vrl1 is a predicted PX-BAR protein that interacts with conserved machinery at the endosome.
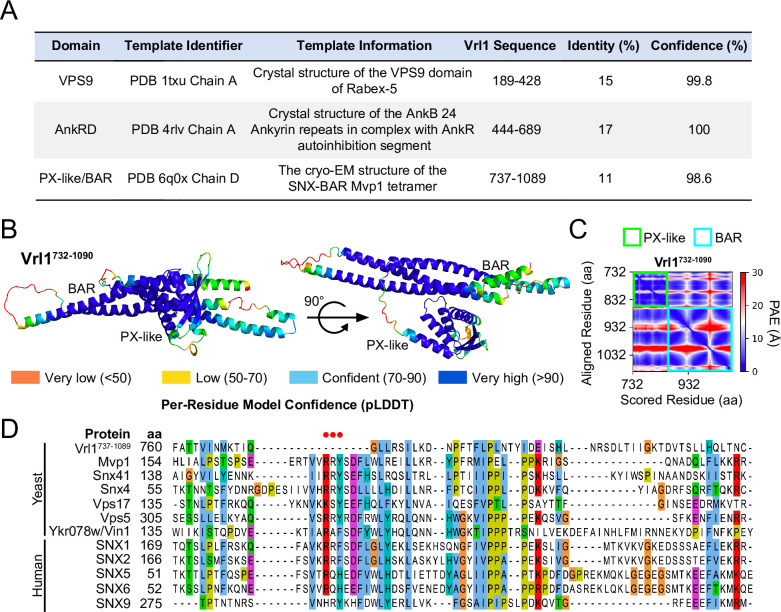
(A) Schematic of Vrl1 and VARP domain architecture. (B) ColabFold predicts the Vrl1 C-terminus has a SNX-BAR-like PX and BAR domain fold. (C) Vrl1-Envy colocalizes with Did2-mRuby2-labeled endosomes, but not with the Sec7-dsRed Golgi marker. (D) Quantification of colocalization as the percentage of Vrl1 puncta overlapping RFP puncta in C. Two-tailed equal variance t test; n=3, cells/strain/replicate ≥1395; ****=p < 0.0001. (E) Schematic of DHFR proximity screen methodology. (F) Z-score distribution of the ratio of colony areas from genome-wide DHFR screens of full-length and truncated Vrl1 baits that localize to the endosome and cytosol, respectively. (G) Gene Ontology (GO) functional enrichment analysis of Vrl1 DHFR interactors (Z-score >2; http://geneontology.org). GO terms of the most specific hierarchical subclass with a fold enrichment value >25 are presented as the negative base 10 log of the associated p-value from a Bonferroni-corrected binomial test of significance. Scale bars, 2 µm. Error bars report standard error of the mean (SEM). Enrich., enrichment. aa, amino acids.