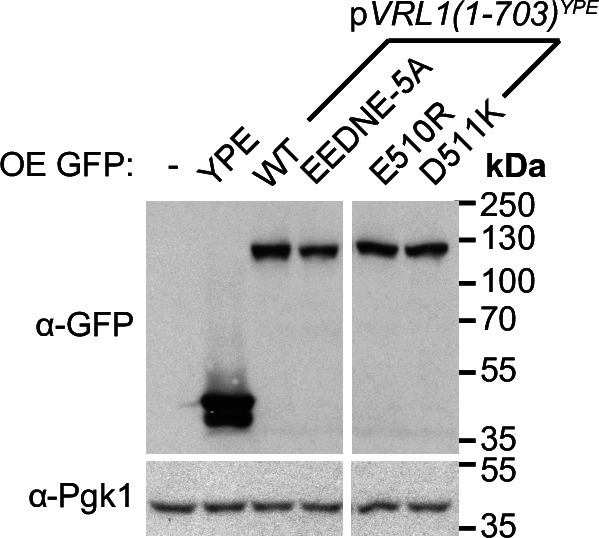
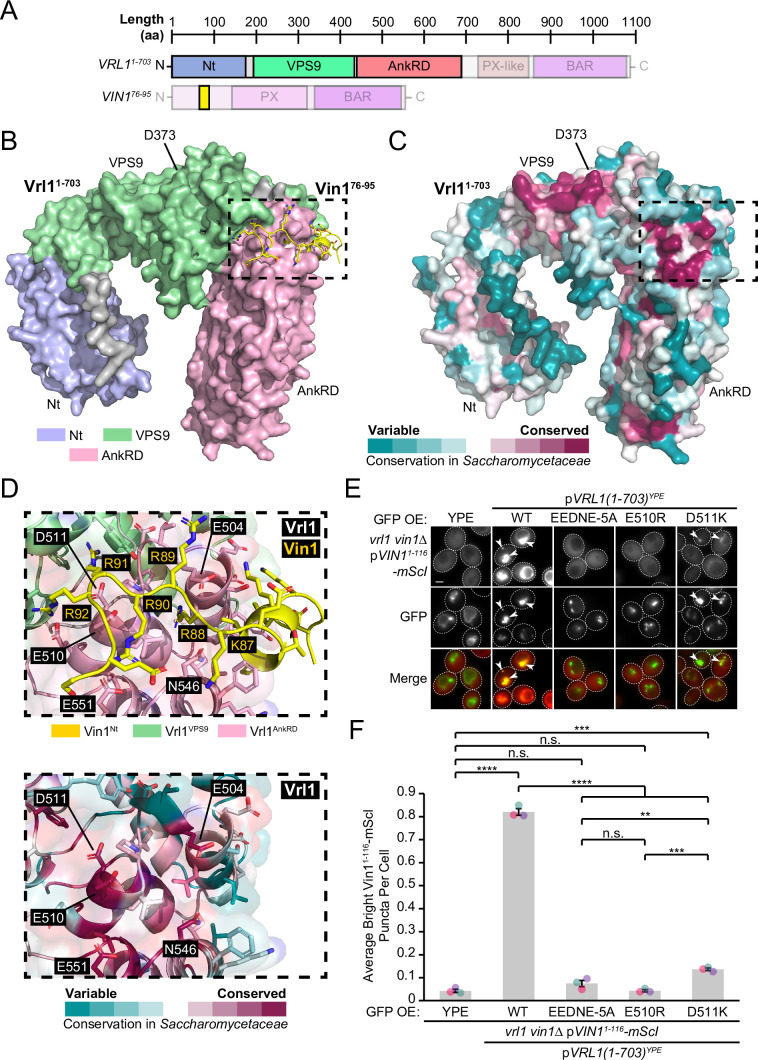
Figure 5. The Vrl1 AnkRD associates with the Vin1 N-terminus through electrostatic interactions.
(A) Schematic of query sequences used to predict the interact between Vrl1 and the Vin1 N-terminus. Modelled regions are shown as completely opaque. (B) ColabFold-predicted interaction between the Vrl1 AnkRD and a minimal fragment of the Vin1 N-terminus (Vin176-95; pTMscore = 0.73). (C) Vrl1 sequence conservation within family Saccharomycetaceae determined by ConSurf and mapped to a surface model that was predicted by ColabFold. Strong sequence conservation can be seen at the predicted Vin176-95 interacting site and near the catalytic D373 residue. (D) Top: Vin176-95 is predicted to associate with Vrl1 through a run of basic residues. Bottom: Acidic and polar residues in the predicted Vin1-associating Vrl1 AnkRD site are among the most conserved within family Saccharomycetaceae. (E) Mutation of acidic and polar residues in the Vrl1 AnkRD reduces recruitment of the Vin1 N-terminus by the Vrl1(1-703)YPE chimera. (F) Quantification of Vin11-116-mScI puncta per cell in E. One-way ANOVA with Tukey’s multiple comparison test; n=3, cells/strain/replicate ≥863; not significant, n.s.=p > 0.05, *=p < 0.05, ***=p < 0.001, ****=p < 0.0001. Scale bars, 2 µm. Error bars report SEM. OE, over-expressed. Nt, N-terminus. WT, wild type. aa, amino acids. YPE, Ypt35(PX)-Envy.
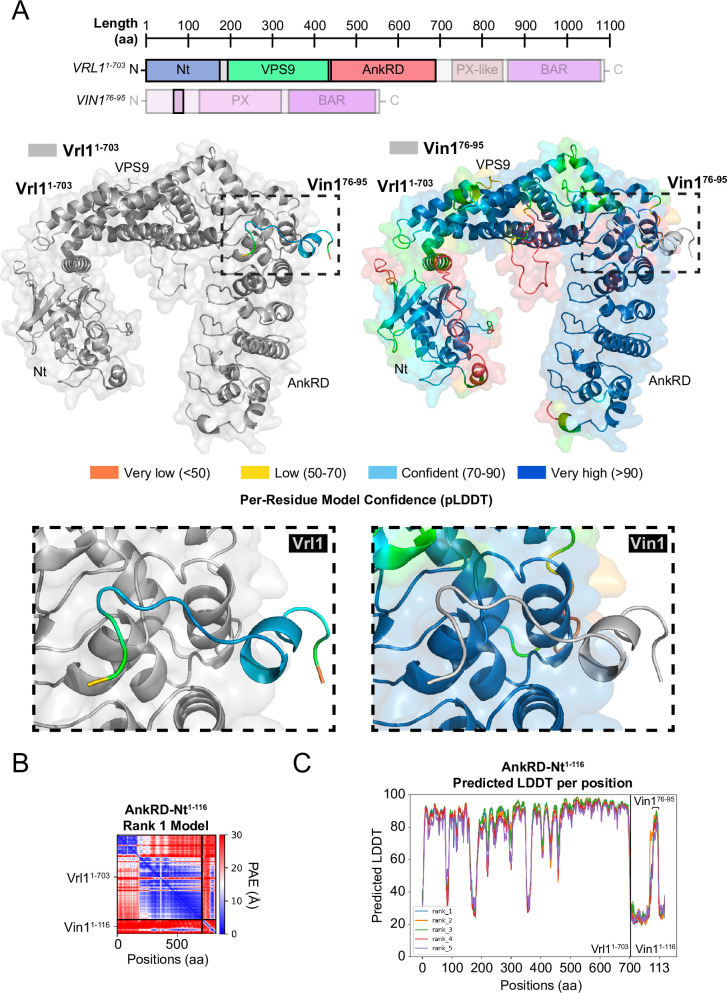
Figure 5—figure supplement 1. Confidence measures of Vrl1 AnkRD-Vin1 N-terminus binding predictions.
Figure 5—figure supplement 2. Vrl1(1-703)YPE chimeras with AnkRD mutations are stably expressed.