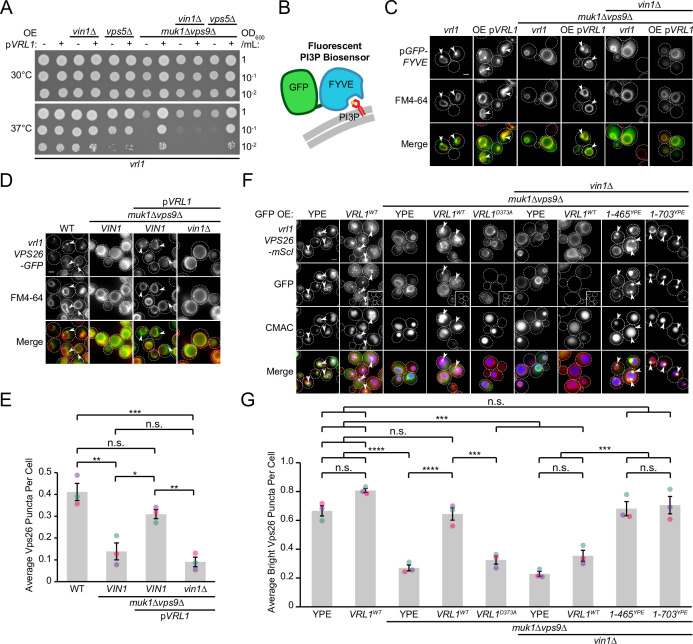
Figure 6. Vin1 controls Vrl1 GEF activity via membrane localization.
(A) Deletion of VIN1, but not VPS5, prevents Vrl1 from rescuing the temperature sensitivity of a strain lacking other VPS9-domain GEFs. (B) Schematic of PI3P-binding fluorescent biosensor. (C) Deletion of VIN1 prevents Vrl1 from stimulating endosomal PI3P production in a strain lacking other VPS9-domain GEFs. (D) Deletion of VIN1 prevents Vrl1 from rescuing Vps26-GFP localization in a strain lacking other VPS9-domain GEFs. (E) Quantification of Vps26-GFP puncta per cell in D. One-way ANOVA with Tukey’s multiple comparison test; n=3, cells/strain/replicate ≥1503; not significant, n.s.=p > 0.05, *=p < 0.05, **=p < 0.01, ***=p < 0.001. (F) Vin1 is dispensable for Vrl1 activity when fragments containing the N-terminus and VPS9 domain are artificially recruited by a YPE endosomal anchor. Insets are scaled to match other images in the same channel (see materials and methods for details). (G) Quantification of Vps26-mScI puncta per cell in F. One-way ANOVA with Tukey’s multiple comparison test; n=3, cells/strain/replicate ≥750; not significant, n.s.=p > 0.05, ***=p < 0.001, ****=p < 0.0001. Scale bars, 2 µm. Error bars report SEM. OE, over-expressed. WT, wild type. YPE, Ypt35(PX)-Envy.