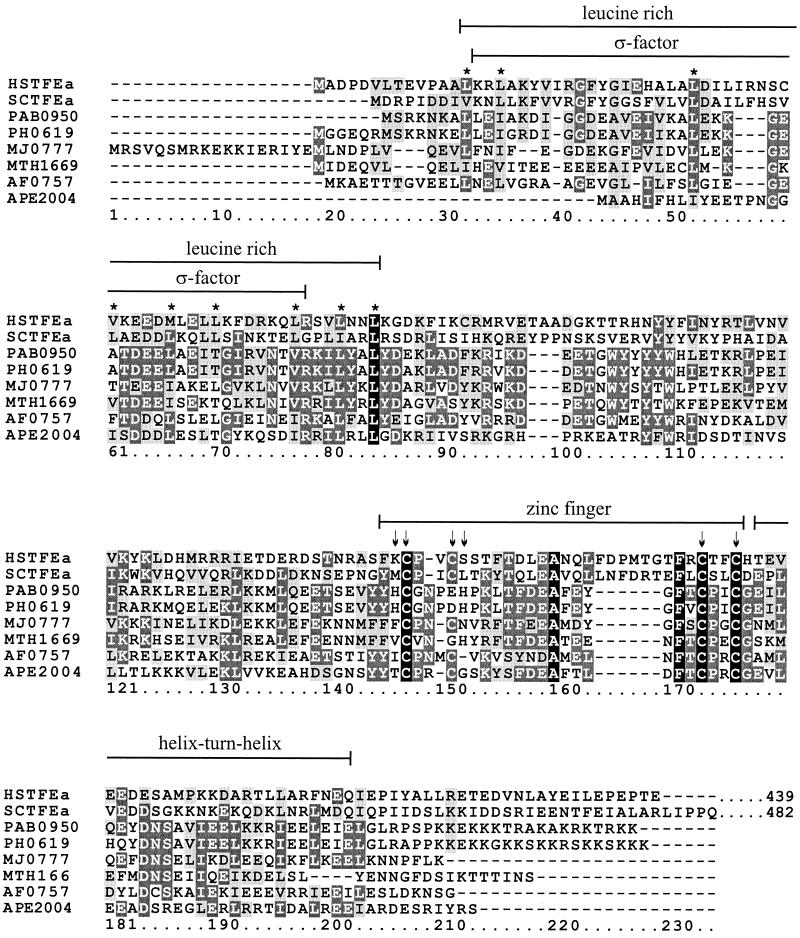
FIG. 1.
Alignment of archaeal sequences with the N-terminal sequences of the α-subunits of yeast and human TFIIE. The M. thermoautotrophicum ΔH (MTH1669), Pyrococcus horikoshii (PH0619), Pyrococcus abysii (PAB0950), Methanococcus jannaschii (MJ0777), Archaeoglobus fulgidus (AF0757), and Aeropyrum pernix (APE2004) archaeal sequences are identified by their genome reading frame reference numbers (2, 15), and the yeast and human sequences are identified by SCTFEa (GenBank accession no. 607957) and HSTFEa (GenBank accession no. 5031726), respectively. Only the N-terminal 210 residues of the yeast (482 residues in total) and human (439 residues in total) proteins are shown. The alignment was generated by PILEUP (Genetic Computer Group, Inc.). Residues present in all eight sequences are identified by a black background, identical residues are identified by a dark gray background, and similar residues are identified by a light gray background. Brackets above the sequences indicate the leucine-rich region with similarity to bacterial ς factors (25), the zinc finger motif (18), and the helix-turn-helix region in human TFIIE. Conserved hydrophobic positions within the leucine-rich region are identified by asterisks (∗), and arrows (↓) identify cysteine and histidine residues that could ligate the zinc atom in the proposed zinc finger domains.