Table 1.
Results from univariate analysis of malaria disease in Afghanistan 2009 with standard errors
| Components | Model 1a (without autoregression) | Model 1b (with fixed parameters only) | Model 1c (with random effects) | Model 1d (with random effects + environmental data) |
|---|---|---|---|---|
| Endemic υi,t | ||||
| α0 (s.e.) | 9·223 (0·493) | 14·532 (5·435) | ||
| Trend α1 (s.e.) | −0·045 (0·079) | 0·254 (0·571) | −0·147 (0·076) | −0·130 (0·075) |
| Sin α2 (s.e.) | 0·923 (0·207) | 6·228 (1·409) | 1·399 (0·201) | 1·330 (0·213) |
| Cos α3 (s.e.) | −1·806 (0·150) | −0·887 (0·438) | −1·621 (0·097) | −2·159 (0·082) |
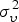 |
2·152 | 2·1231 | ||
| IID random effects | 8·302 (0·485) | 8·419 (0·506) | ||
| Autoregressive λi,t | ||||
| β0 (s.e.) | −0·031 (0·037) | −0·518 (0·100) | −1·165 (0·931) | |
| Log Alt β1 (s.e.) | 0·077 (0·122) | |||
| Precip β2 (s.e.) | 0·015 (0·005) | |||
| Temp β3 (s.e.) | 1·125 (0·022) | |||
| Neighbour-driven ϕ | ||||
| γ0 (s.e.) | −6·269 (0·906) | −5·664 (0·435) | −5·488 (0·526) | −5·610 (0·660) |
| Overdispersion ψ (s.e.) | 1·425 (0·116) | 0·188 (0·019) | 0·112 (0·012) | 0·111 (0·012) |
| AIC | 3248·06 | 2740·09 | ||
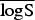 (P values) (P values) |
7·169 (0·0091) | 6·653 (0·0440) | 6·815 (0·0739) | 6·763 |
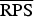 (P values) (P values) |
509·359 (0·0007) | 209·019 (0·0011) | 133·606 (0·0012) | 117·505 |
IID, Independent and identically distributed; AIC, Akaike's Information Criterion.
In the rows for 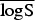 and
and 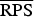 measures, the Monte Carlo P values give comparisons of malaria models 1a–1c to the best model (model 1d) based on permutation tests for paired observations (9999 permutations).
measures, the Monte Carlo P values give comparisons of malaria models 1a–1c to the best model (model 1d) based on permutation tests for paired observations (9999 permutations).
