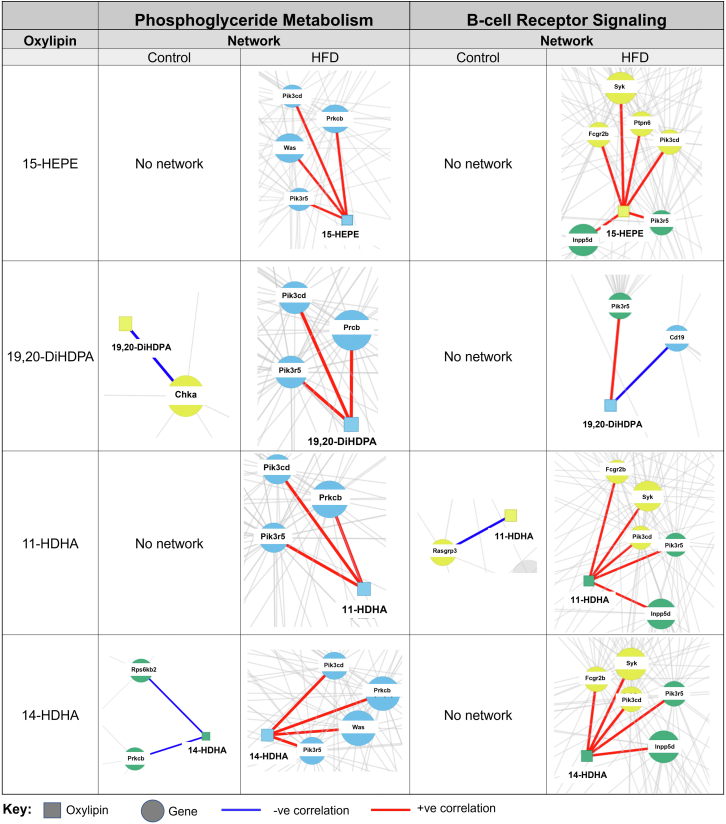
Figure 6.
Integrative omics of transcriptomic and metabolomic analyses shows different network interactions for key n-3 PUFA-derived oxylipins with a high fat diet. Differential network analysis for selective n-3 PUFA-derived oxylipins (15-HEPE, 19,20-DiHDPA, 11-HDHA, and 14-HDHA) was conducted for genes present in the Phosphoglyceride metabolism and B-cell receptor signaling pathways using xMWAS, a software program for data integration. Squares represent metabolites and circles represent genes. Clustering of data resulted in distinct communities which are represented by different colors for genes and metabolites. Blue lines represent a negative correlation and red lines represent a positive correlation between a given oxylipin and gene.