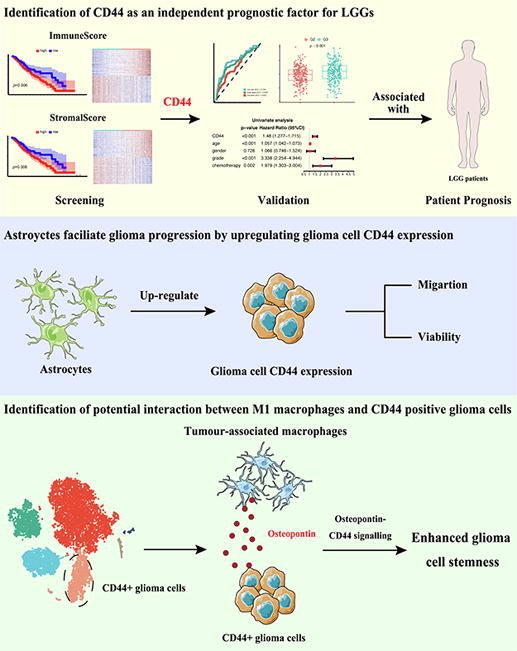
Graphical abstract
Keywords: CD44, Tumour microenvironment, Lower-grade gliomas, Prognosis, Glial cells
Abbreviations: LGGs, Lower-grade gliomas; TME, Tumour microenvironment; GBM, Glioblastoma; CSCs, Cancer stem cells; GSCs, Glioma stem cells; OPN, Osteopontin; SPP1, Secreted phosphoprotein 1; TAMs, Tumour-associated macrophages; TIC, Tumour-infiltrating immune cell; TCGA, The Cancer Genome Atlas; CGGA, The Chinese Glioma Genome Atlas; ACM, Astrocyte-conditioned medium; PDL, Poly-d-lysine; OS, Overall survival; BBB, Blood–brain barrier; RT-qPCR, Real-time quantitative polymerase chain reaction
Abstract
Because of the heterogeneity of lower-grade gliomas (LGGs), patients show various survival outcomes that are not reliably predicted by histological classification. The tumour microenvironment (TME) contributes to the initiation and progression of brain LGGs. Identifying potential prognostic markers based on the immune and stromal components in the TME will provide new insights into the dynamic modulation of these two components of the TME in LGGs. We applied ESTIMATE to calculate the ratio of immune and stromal components from The Cancer Genome Atlas database. After combined differential gene expression analysis, protein–protein interaction network construction and survival analysis, CD44 was screened as an independent prognostic factor and subsequently validated utilizing data from the Chinese Glioma Genome Atlas database. To decipher the association of glioma cell CD44 expression with stromal cells in the TME and tumour progression, RT–qPCR, cell viability and wound healing assays were employed to determine whether astrocytes enhance glioma cell viability and migration by upregulating CD44 expression. Surprisingly, M1 macrophages were identified as positively correlated with CD44 expression by CIBERSORT analysis. CD44+ glioma cells were further suggested to interact with microglia-derived macrophages (M1 phenotype) via osteopontin signalling on the basis of single-cell sequencing data. Overall, we found that astrocytes could elevate the CD44 expression level of glioma cells, enhancing the recruitment of M1 macrophages that may promote glioma stemness via osteopontin-CD44 signalling. Thus, glioma CD44 expression might coordinate with glial activities in the TME and serve as a potential therapeutic target and prognostic marker for LGGs.
1. Introduction
Diffuse gliomas are the most common malignant brain tumours in adults, and they also have the highest mortality and recurrence rate [1]. Based on histology and clinical criteria, gliomas are classified as Grades I to IV [2]. Accordingly, WHO grade II and III gliomas are termed diffuse lower‐grade gliomas (LGGs), which are intracranial malignant tumours that originate from astrocytes or oligodendrocytes alone or oligo-astrocytomas (mixed astrocytes and oligodendrocytes) [3], [4]. They often arise in young patients and have an indolent course, with longer survival than glioblastoma (GBM, WHO grade IV) [5]. The survival of LGG patients ranges from 1 to 15 years [1]. Therefore, unlike glioblastoma, LGG often affects the health of patients in a chronic and long-term manner. Despite recent efforts to improve the prognosis of patients with LGGs, half of LGG patients present with highly aggressive, drug-resistant gliomas [4]. Because of this aggressive behavior, LGGs cannot be completely cured and always recur and progress to GBM [6]. Thus, delaying tumour onset and reducing tumour progression are the most challenging issues. Radiotherapy and chemotherapy, however, fail to markedly prolong patient survival [7], [8]. Therefore, we sought to identify novel biomarkers to predict glioma prognosis and guide treatment, improving the prognosis and quality of life of LGG patients.
Therapeutic resistance inevitably develops despite effective targeted treatment of tumours [9]. Research dissecting the mechanisms of therapeutic resistance identified the importance of the tumour microenvironment (TME) during tumour progression, whereby the TME, composed of noncancerous cells, tumour vasculature, and extracellular matrix, coevolves with neoplastic cells to achieve malignancy development [10]. These extrinsic compartments of tumour cells take part in dictating anomalous cellular functions and exacerbating the development of tumour malignancies [11]. Recent studies have implied that cancer stem cells (CSCs) are associated with poor prognosis in tumour patients [12]. CSCs are characterized as a subpopulation of tumour cells with properties such as the ability to self-renew, undergo unlimited growth, high capacity of tumour initiation and maintain the stem of tumour growth [13]. Efforts have been made to clarify whether CSCs contribute to GBM tumour initiation, progression, relapse, and resistance to treatment [14]. The TME stimulates CSC self-renewal and tumorigenicity to aggravate the downstream effects [15]. Molecular markers associated with glioma stem cells (GSCs) include CD133, CD44, SOX2, and STAT3 [16]. CD44 is involved in the mesenchymal characteristics of GBM cells, such as cell invasion and therapeutic resistance [17]. Pietras and collaborators proposed that astrocytes enhance glioma stem cell phenotypes via activation of CD44 signalling [18]. In addition, CD44 expression is also found in tumour microenvironmental cells. In the TME, the CD44 expression of GFAP positive astrocytes like cells was shown to be different between LGGs and GBM [19]. As CD44 is also a panreactive marker for reactive astrocytes, there may be a coordinated regulation of CD44 expression between astrocytes and glioma cells during glioma development [20].
Osteopontin (OPN), also known as secreted phosphoprotein 1 (SPP1), is one of the classical ligands of CD44 [21]. Numerous studies have suggested that SPP1 expression is negatively correlated with glioma patient survival [22], [23], [24]. It has been suggested that OPN-CD44 signalling in the glioma perivascular niche plays a role in promoting cancer stem cell phenotypes and tumour growth [18]. A wide variety of cells secrete OPN to promote tumour growth, especially tumour-associated macrophages (TAMs) [25]. TAMs constitute up to 50 % of live cells in GBM tumours [26]. Instead of inhibiting tumour growth, TAMs secrete OPN to facilitate glioma invasion and the formation of an immunosuppressive microenvironment, leading to aggressive tumour growth [22], [23], [27]. Macrophage polarization has significant effects on macrophage functions, which can be divided into the M1 (proinflammatory/tumour resistance) and M2 (anti-inflammatory/tumour promotion) states [28]. These abundant stromal cells have been an emerging target for glioma therapy [29]. The symbiotic interplay between glioma and glial cells sustains the survival of glioma cells and accelerates tumour progression. However, existing research on the TME of LGGs is still deficient, let alone the interaction of glioma and glial dynamics in the TME. Thus, further studies on the TME of LGGs are needed to predict prognosis and provide solid evidence for novel therapeutic targets.
In this study, we applied the ESTIMATE [30] and CIBERSORT [31] computational methods to calculate the ratio of immune and stromal components and the tumour-infiltrating immune cell (TIC) proportion in The Cancer Genome Atlas (TCGA) database. Through PPI network construction and Cox regression analysis, together with The Chinese Glioma Genome Atlas (CGGA) database, CD44 was comprehensively identified as a predictive biomarker for LGGs. In addition, survival analysis, clinal correlations and GO and KEGG enrichment analyses were performed to collectively determine the potential pathways and mechanisms related to CD44. Our experiments implied that astrocytes may enhance migration and cell viability in the glioma cell line U87 by upregulating glioma CD44 expression. Surprisingly, further analysis indicated that CD44 expression was positively associated with M1 macrophage infiltration and that CD44+ glioma cells might interact with M1 macrophages via OPN signalling. Together, these findings suggest an association between glioma CD44 expression and glial cell dynamics in the TME during glioma progression.
2. Materials and methods
2.1. Data acquisition
Transcriptome RNA sequencing data of 529 LGG cases from the TCGA cohort and relevant clinical data were downloaded from UCSC Xena Browser (version 07–19-2019) [33]. Transcriptome RNA sequencing and clinical data of 1018 glioma cases from 2 cohorts, CGGA_325 (referred to as the CGGAseq1 cohort) and CGGA_693 (referred to as the CGGAseq2 cohort), were downloaded from the CGGA database (https://www.cgga.org.cn). Transcriptome profiles of 20 normal tissue (nonglioma) samples were also downloaded from the CGGA database (referred to as the CGGAnormal cohort).
2.2. Generation of the ImmuneScore, StromalScore, and ESTIMATEScore
ESTIMATE was used to evaluate the proportions of immune and stromal components in the TME of samples [30]. For each sample, ImmuneScore, StromalScore and ESTIMATEScore were reported, indicating the relative proportions of immune components and stromal components and the sum of them, respectively. A higher score suggests a larger proportion of the corresponding component in the TME.
2.3. Survival analysis
Cases with detailed records of overall survival time longer than 30 days (1 month) were retained for analysis. A total of 477, 170, and 420 cases were retained from the TCGA, CGGAseq1 and CGGAseq2 cohorts, respectively. The R package survival was used to conduct survival analysis. The Kaplan–Meier method was employed to plot the survival curve, and the log rank test was used for statistical significance testing. Univariate Cox regression and multivariate Cox regression were performed to assess the relationship between survival and gene expression, along with clinicopathological characteristics. The hazard ratio and statistical significance determined by the Wald test were reported for each feature. The specificity and sensitivity of using CD44 expression to predict survival for one-year, three-year and five-year spans were determined using ROC curve analysis utilizing the R package survivalROC [34].
2.4. Generation of Differentially expressed genes
The R package limma was employed for gene differential expression analysis. Differentially expressed genes (DEGs) with an absolute fold change larger than 1.5 and a false discovery rate (FDR) < 0.05 were considered to be statistically significant [35]. When performing DEG analysis between tumour and normal tissue samples, to increase the statistical power by increasing the sample size, the transcriptome profiles of tumour samples from the two CGGA cohorts were integrated by ComBat [36].
2.5. Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed utilizing the GSEA function implemented in the R package clusterProfiler [37] and using the reference gene sets of hallmark gene sets (H), canonical pathways of curated gene sets (C2), and biological processes and molecular functions of ontology gene sets (C5:GO) from the MSigDB database [38]. An adjusted p value < 0.05 was considered to indicate significant enrichment.
2.6. Protein–Protein interaction network construction
A protein–protein interaction (PPI) network was constructed with Cytoscape [39] version 3.9.0, utilizing PPI information from the STRING database [32]. Nodes with confidence of interactive relationship greater than 0.95 were used to establish the network. Only the largest subnetwork was retained to exclude unconnected subnetworks. Utilizing the Cytoscape plugin CytoHubba [40], six measurements based on the shortest paths, including closeness, eccentricity, radiality, bottleneck, stress and betweenness, were calculated for node ranking to determine the important nodes (i.e., hub genes). The genes in the intersection set of the top 15 ranked nodes of each measurement were defined as hub genes.
2.7. Immunohistochemical staining
Immunohistochemical staining images of CD44 protein in normal, LGG and GBM tissues were obtained from the HPA (https://www.proteinatlas.org/) to determine the differences in CD44 expression at the protein level. The antibody HPA005785 was used for IHC.
2.8. Tumour-Infiltrating immune cell profile
To determine the tumour-infiltrating immune cell (TIC) abundance profile for all tumour samples, CIBERSORT was used [31]. After quality filtering with p < 0.05, 125 tumour samples were retained for further analysis.
2.9. Cell culture and preparation of conditioned medium
Trans serum-free primary astrocyte cultures were prepared as described previously with modifications [41], [42]. Briefly, cortices from 1 to 3 postnatal C57BL/6 mice were dissected and rinsed in cold Hank’s balanced salt solution (HBSS), and the meninges were carefully removed. Tissue was mechanically dissociated and triturated to produce single cells. After centrifugation, the pellet was resuspended in serum-containing medium (DMEM containing 10 % foetal bovine serum (FBS) and 0.5 % penicillin/streptomycin). Cells were plated in T-75 flasks previously coated with poly-d-lysine (PDL, Sigma) and incubated at 37˚C in a humidified 5 % CO2, 95 % air chamber. After incubation for approximately 5–7 d until confluence was reached, the cells were shaken for 9 h at 200 rotations per min (RPM) on an orbital shaker to remove endothelial cells, microglia and oligodendrocyte precursor cells. Purified astrocytes were plated onto PDL–coated 60 mm dishes at 500,000 cells. Subsequently, trans serum-free astrocytes were cultured with serum-free medium containing 50 % neurobasal, 50 % DMEM, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 292 μg/ml l-glutamine, 1 × SATO, 5 μg/ml N-acetyl cysteine and 5 ng/ml HBEGF (Peprotech, 100–47). Then, half of the medium was changed every day. Astrocytes cultured in serum-containing medium were continuously cultured in the same medium. U87 glioma cell lines were obtained from American Type Culture Collection (ATCC). Cells were cultured in cell culture dishes with DMEM containing 10 % FBS. To prepare the astrocyte-conditioned medium (ACM), when astrocytes were cultured in serum-free medium for 3 days, the supernatants were collected and centrifuged at 3000 × g for 3 min and stored at − 80 °C until use.
2.10. Immunofluorescence
Cells were seeded at 6 × 104 cells/well in 12-well plates on PDL coated coverslips in serum free or serum-containing medium. Astrocytes were fixed in 4 % paraformaldehyde, blocked with 5 % NDS in 0.3 % PBT and 0.1 % NaN3 for 20 min, and immunostained with glial fibrillary acidic protein (GFAP, Abcam, ab53554), diluted 1:250, overnight at 4 °C. Donkey anti-goat IgG (H + L) cross-adsorbed secondary antibody, Alexa Fluor 555 (A21432, Invitrogen), diluted 1:250 were applied for 30 min at room temperature. Images were taken at 40X magnification on a Leica DM6 microscope.
2.11. Quantitative Real-Time PCR
An RNA-Quick Purification Kit (Yishan, Shanghai, China) was used to extract the total RNA of cells. With PrimeScript 1st Strand complementary DNA Synthesis kit (Takara, Dalian, China), 1 μg of RNA was reverse transcribed into cDNA in a final volume of 20 μL. Real-time PCRs were conducted with a PrimeScript® miRNA RT–PCR Kit (Takara, Dalian, China) and a Real-Time PCR System (CFX Connect, BIORAD) in line with the manufacturer’s protocols. Actin served as an internal reference. RT-qPCR was performed with primer pairs for: CD44: forward (5′-CTGCCGCTTTGCAGGTGTA-3′) and reverse (5′-CATTGTGGGCAAGGTGCTATT-3′) and for Actin as a control: forward (5′-TCAAGATCATTGCTCCTCCTGAG-3′) and reverse (5′-ACATCTGCTGGAAGGTGGACA-3′).
2.12. Wound healing assays
U87 cells (5 × 105 cells) were plated onto a 12-well plate in serum-containing medium. After incubation for 1–2 days until U87 cells were confluent, cells were wounded with a plastic pipette tip. Dead cells were removed by rinsing twice with phosphate-buffered saline (PBS). U87 cell cultures were subjected to two experimental groups: (1) Control: U87 cells cultured in serum-free medium; (2) U87 + ACM: U87 cells cultured in astrocytes conditioned medium (ACM). Once serum-free medium or ACM was added, and the cells were incubated in a chamber. At various timepoints (0 h, 12 h, 24 h, 48 h, and 72 h), images of the lesion border were acquired using a Motic inverted microscope (AE31 EF-INV). The wound healing ability was calculated based on the captured images.
2.13. Cell viability assays
U87 cells (8,000 cells) were plated onto a 96-well plate in serum-free medium or ACM. After incubation for 24 h, 10 μL of CCK8 (Beyotime, C0037, China) was added to each well. The optical density value was measured at 450 nm after 2 h.
2.14. Single-cell RNA sequencing data analysis
The processed per-cell read count matrices of 10 astrocytoma samples were obtained from the GEO database (dataset accession numbers GSE89567 and GSE138794, Supplemental Table 5). Analysis was performed using the Seurat pipeline [43]. For dataset GSE138794, in which the gene expression was obtained by the 10X droplet-based platform, low-quality cells were filtered by thresholding the number of detected genes<200 or more than 6000, detected molecules<1000 reads, or mitochondrial gene expression higher than 15 % in the cells. For dataset GSE89567, in which the gene expression was obtained by the SMART-seq2 platform, TPM-normalized expression values were obtained and then transformed to use the same scale factor. Batch effects between two datasets and among samples were corrected by Harmony [44] for integrated analysis. The significant DEGs of each cell cluster were obtained by the Wilcoxon two-tailed test. To annotate the cell type of each cell cluster, the DEGs of each cell cluster were intersected with canonical markers of different cell types: oligodendrocytes (MBP, MOBP, PLLP, CLDN11, PLP1), malignant-astrocyte-like cells (GFAP, ALDOC, HEY1, ID3, ID4), macrophages (CD14, IFNGR1, C1QB, CSF1R, HLA-DRA, AIF1), malignant-stem-like cells (CCND2, PDGFRA, SOX4), fibroblasts (ACTA2, ADIRF, VIM, TAGLN) and proliferating cells (HMGB2, GMNN, TYMS). To further confirm the malignancy status of the two annotated malignant cell clusters, single-cell copy number variation (CNV) analysis was performed using CopyKAT [45]. To identify subclusters among macrophages, macrophages were extracted, clustered and further annotated according to their cell origins and inflammation-related gene expression. Differentially expressed genes and corresponding fold changes for GSEA were obtained for the two macrophage subclusters. The gene module expression scores were calculated using the AddModuleScore function in Seurat to indicate the cell origins and biological effects of macrophages. The cell interactions between different cell types were inferred using CellPhoneDB [46] (sample MGH103 was excluded from this analysis because no macrophages were detected in the sample).
2.15. Statistical analysis
All statistical analyses in this study were conducted using R software (version 4.0.3) and GraphPad Prism (version 8.0.2). The Wilcoxon rank-sum test was used to evaluate statistical significance when comparing two groups. Comparisons among three or more groups were performed using Kruskal–Wallis tests. A two-tailed p value of < 0.05 was considered statistically significant unless otherwise indicated. Multiple testing correction was performed using the Benjamini–Hochberg method when applicable.
3. Results
3.1. Correlation of scores with the survival and clinicopathological stage of LGG patients
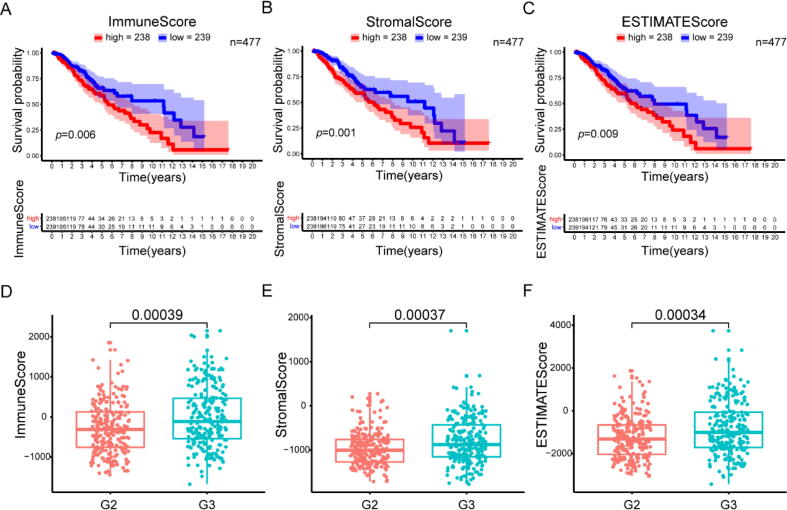
To determine the correlation of the ratio of immune and stromal components with the survival of LGG patients, we employed the ImmuneScore, StromalScore, and ESTIMATEScore to conduct Kaplan–Meier survival analysis in the TCGA database. The higher scores estimated for the ImmuneScore and StromalScore suggest a larger extent of the immune and stromal components in the TME, which could be represented by the higher score estimated for the ESTIMATEScore. The respective proportions of immune and stromal components were negatively correlated with the overall survival (OS) rate (Fig. 1A, B). Similarly, the ESTIMATEScore had a significant negative correlation with the OS rate (Fig. 1C). Thus, immune and stromal components in the TME are both valuable for predicting the clinical outcomes of LGG patients. Subsequently, to identify the association between the estimated proportion of immune and stromal components and the clinicopathological stage, the grade of LGG cases was analysed with the ImmuneScore and StromalScore. Displayed in Fig. 1, the ImmuneScore and StromalScore increased significantly with increasing tumour grade, suggesting that the ImmuneScore and StromalScore had a positive correlation with the grade of LGGs (Fig. 1D, E). In addition, the ESTIMATEScore increased significantly with progression of the stage of LGGs (Fig. 1F). However, neither score was correlated with age or sex (Fig. S1, Supplement Table 1). These results imply that the proportions of immune and stromal components are associated with the survival and progression of LGGs.
Fig. 1.
Scores were correlated with the survival and clinicopathological staging characteristics of LGG patients. Kaplan–Meier survival analysis of LGG patients labelled with high or low scores for (A) ImmuneScore (p = 0.006), (B) StromalScore (p = 0.001) and (C) ESTIMATEScore (p = 0.009) compared with the median. The correlation between (D) ImmuneScore (p = 0.00039), (E) StromalScore (p = 0.00037) and (F) ESTIMATScore (p = 0.00034) and stage.
3.2. The differences in Immune-related genes were underlined by DEGs shared by ImmuneScore and StromalScore
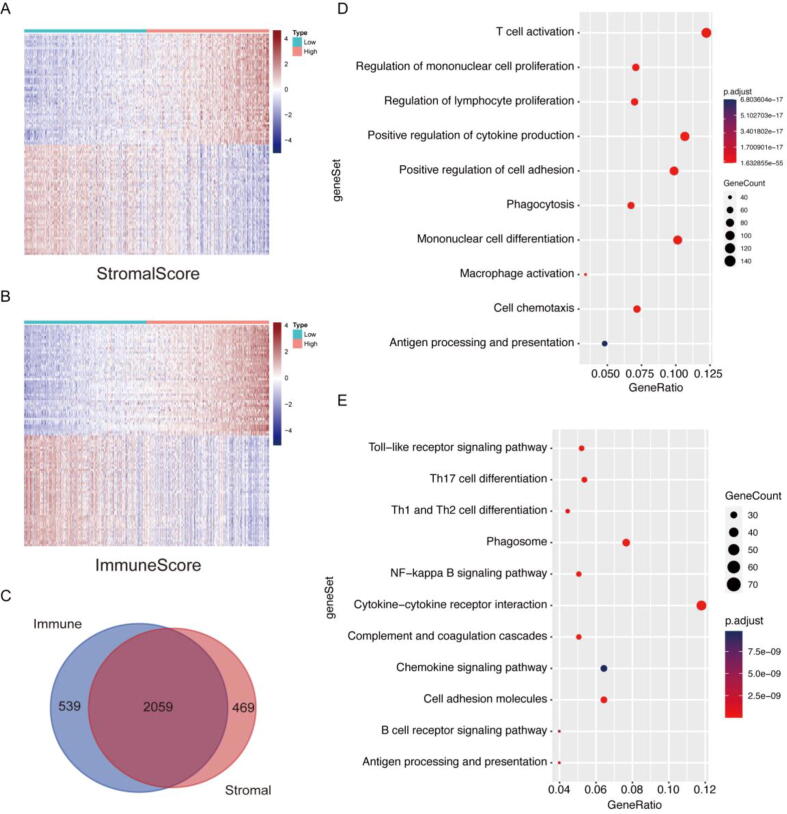
As a high ratio of immune and stromal components in the TME of LGG patients might lead to worse prognosis, the differences in gene expression profiles between LGG patients with different ratios of immune and stromal components in the TME were investigated to decipher the TME-related gene signature. High- and low-score samples were analysed to obtain DEGs (Fig. 2A, B). On the one hand, from ImmuneScore, 2598 DEGs were identified, and among them, 1387 genes were upregulated in comparison with the median (samples with high score vs low score, Supplement Table 3). On the other hand, from StromalScore, we obtained 2528 DEGs, which included 1559 upregulated genes (Supplement Table 4). As shown in the Venn diagram, a total of 2059 DEGs shared by ImmuneScore and StromalScore were identified through intersection analysis (Fig. 2C). These DEGs may have an important impact on the status of the TME. Therefore, 1258 upregulated genes shared by the high-ImmuneScore group and high-StromalScore group were used in ClusterProfiler to determine their functional significance. Gene ontology (GO) enrichment analysis showed that these DEGs mapped to immune-related GO terms, such as T-cell activation, regulation of mononuclear cell proliferation and macrophage activation (Fig. 2D). The results from Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis confirmed the enrichment of the Toll-like receptor signalling pathway, NF-kappa B signalling pathway and B-cell receptor signalling pathway (Fig. 2E). These data suggest that the upregulated DEGs shared by ImmuneScore and StromalScore map to immune-related activities. Higher scores implied enhanced immune activities in the TME. As a result, the ratio of immune and stromal components is positively correlated with the immune activities in the TME, indicating the significant alteration of immune activities in the TME across LGG patients with different ratios of immune and stromal components.
Fig. 2.
Heatmap, Venn diagrams and enrichment analysis of DEGs shared by StromalScore and ImmuneScore. (A) Heatmap of the top 50 upregulated and downregulated DEGs for the StromalScore, generated by comparing the high score group with the low score group. (B) Heatmap of the top 50 upregulated and downregulated DEGs for the ImmuneScore. (C) Venn diagram showing the DEGs shared by ImmueScores and StromalScores. (D) GO enrichment analysis of 1258 shared upregulated genes. (E) KEGG enrichment analysis of shared upregulated genes.
3.3. CD44 was identified as an important factor by analysing the TCGA and CGGA databases
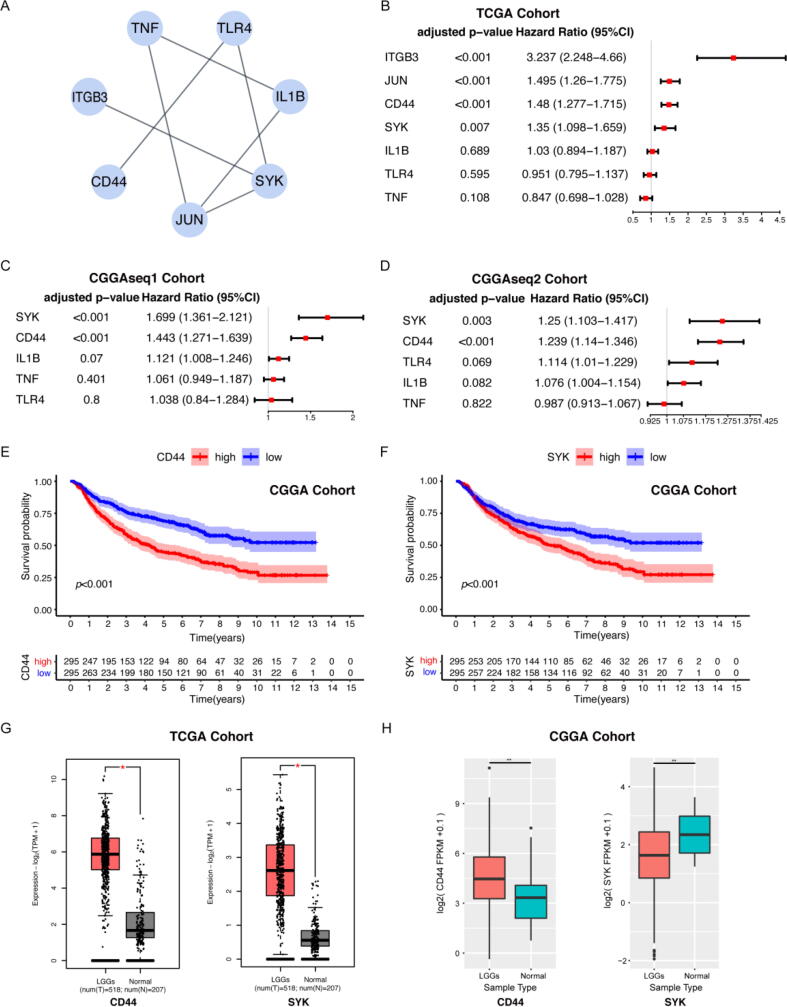
Given that the scores were associated with immune activities in the TME, we next sought to determine the key indicators of TME immune modulation from the perspectives of the ImmuneScore and StromalScore. Using the data from the TCGA database, a PPI network was constructed using the 1258 upregulated genes detected in both the high-ImmuneScore group and the high-StromalScore group (Figs. S2, S3). As there were still too many genes, to increase the sensitivity and specificity, we utilized CytoHubba to discover important nodes and subsequently acquired 7 hub genes. The interactions between them are shown in Fig. 3A. Univariate Cox regression analysis of the survival of LGG patients was implemented to determine the significant factors among the 2059 DEGs. Among the 7 hub genes, the expression levels of CD44, SYK, JUN and ITGB3 were found to be positively correlated with patient prognosis (Fig. 3B).
Fig. 3.
CD44 was identified as an important factor associated with LGG patient prognosis. (A) Interaction network constructed with the 7 hub genes with an interaction confidence value greater than 0.95. (B) Univariate Cox regression analysis of 7 hub genes, which showed that only 4 factors (including CD44) reached statistical significance. Univariate Cox regression analysis of 5 hub genes in the CGGAseq1 cohort (C) and CGGAseq2 cohort (D). Survival analysis of LGG patients with distinct CD44 expression (E) and SYK expression (F). Patients were grouped into high expression or low expression groups relative to the median expression level. p < 0.001 by log-rank test. Differential expression of CD44 and SYK in normal and tumour samples in the TCGA (G) and CGGA (H) databases. Only the expression of CD44 was much higher in the tumour (red) than in normal tissue (grey and green) in both cohorts, with p < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To select a gene for further analyses, we conducted differential gene expression analysis between LGG and normal tissue samples in the CGGA database. A total of 3346 genes were identified as upregulated genes in tumour samples, and 3859 genes were identified as downregulated genes (Fig. S4). Among the 7 hub genes, CD44 was significantly upregulated in tumours, while SYK, IL1B, TLR4 and TNF were significantly downregulated. We further performed univariate Cox regression for all detected DEGs in the two CGGA cohorts separately. Only the expression levels of CD44 and SYK were positively correlated with patient survival in the CGGA database (Fig. 3C, D, E, F). The differential expression of CD44 and SYK between LGGs and normal tissue samples was validated using the TCGA, CGGA and GTEx databases [47]. The expression of CD44 in the tumour samples was significantly higher than that in the normal samples (Fig. 3G, H). Although higher SYK expression was associated with worse prognosis, SYK was contradictorily downregulated in tumours in the CGGA database (Fig. 3F, H). Therefore, we chose CD44 as the core factor for further study. All LGG samples in the TCGA and CGGA databases were divided into a CD44 high-expression group and a CD44 low-expression group, compared with the CD44 median expression. As shown in the survival analysis, LGG patients with higher CD44 expression had shorter survival than those with low CD44 expression in the TCGA and CGGA databases (Fig. 3E, Fig. S5). These data indicate that CD44 expression is negatively correlated with the prognosis of LGG patients.
3.4. CD44 had independent prognostic value for LGG patients
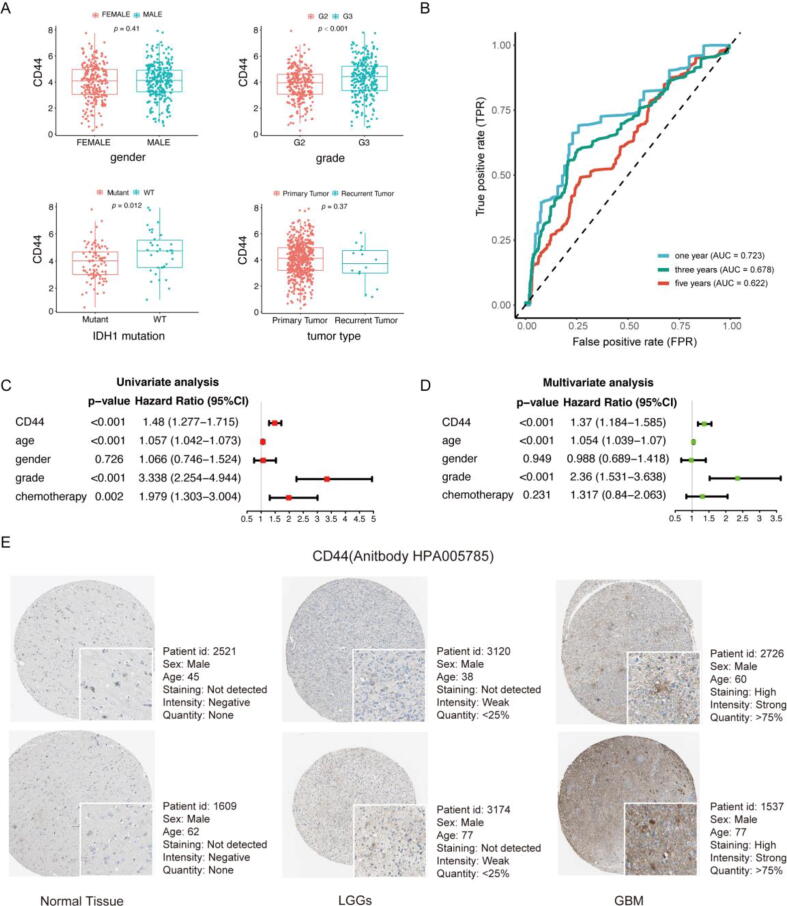
To reveal the association between CD44 expression and the clinicopathological characteristics of LGG patients, data from the TCGA cohort were studied first (Supplement Table 1). The CD44 expression level of LGG patients was associated with grade and IDH mutation (Fig. 4A). The TCGA database lacks more detailed clinical information for different LGG patients. To holistically reveal the relationship between CD44 mRNA expression levels and various clinicopathological characteristics of gliomas, correlations were explored in the CGGA cohorts. The clinicopathological characteristic statistics of the patients are shown in Supplemental Table 2. Under codeletion of 1p19q and IDH mutation, CD44 expression was downregulated compared to noncodeletion and wild-type status, respectively (Fig. S6A, D). In addition, the expression of CD44 was associated with age, chemotherapy, grade and primary/recurrent/secondary-tumour (PRS) type (Fig. S6B, C, E, F). Among various histopathological types of glioma, CD44 expression varied, and CD44 expression reached the highest level in GBM, recurrence of GBM and secondary GBM (Fig. S6G). The results above demonstrate that CD44 expression is associated with the survival and clinicopathological characteristics of LGG patients.
Fig. 4.
CD44 is an independent prognostic factor for LGG patients. (A) The correlation of CD44 expression and the clinicopathological characteristics of LGG patients in the TCGA cohort. (B) The prognostic value of CD44-based prognostic indexes was confirmed by survival-reliant ROC curves. Forest plot displaying the HR with 95% CI of CD44 in glioma patients based on univariable (C) and multivariable (D) analysis. (E) The representative protein expression of CD44 in normal cortex, LGG and GBM tissue. Data were from the Human Protein Atlas (http://www.proteinatlas.org) database.
To verify the prognostic value of CD44 in the TCGA database, we performed univariate and multivariate independent prognostic analyses, and the results showed that high CD44 dexpression, higher grade and advanced age were correlated with shorter survival time of LGG patients (Figure 4C, D). Then, ROC analysis was performed, and the ROC curve is shown in Figure 4B. The area under the curve (AUC) values at one year, three years and five years were all greater than 0.6, which suggested that CD44 had moderate accuracy in predicting the survival of LGG patients (Figure 4B). To validate the detailed prognostic value of CD44 in LGG patients with different clinicopathological characteristics, the correlation between CD44 expression and the prognosis of G2, G3 grade and IDH-mutant LGG patients was further analysed. As expected, CD44 had prognostic value for G2 and G3 grade as well as IDH-mutant LGG patients (Fig. S7A, B, C). Together, these data reveal that CD44 may serve as an independent prognostic factor for LGG patients.
Then, we validated the prognostic value of CD44 in the two validation CGGA cohorts. We conducted a single-gene survival analysis of CD44. We obtained a result similar to that obtained with the TCGA database, and the survival of the CD44 low expression group was longer than that of the high expression group (Fig. S8A, E). Cox regression analysis of the two cohorts also identified CD44 as an essential prognostic factor (Fig. S8C, D, G, H). ROC analysis further validated the reliable prognostic value of CD44. The AUCs of CD44 were 0.725, 0.734 and 0.756 in the CGGAseq1 cohort (Fig. S8B) and 0.549, 0.607 and 0.639 in the CGGAseq2 cohort (Fig. S8F). In addition, the representative protein expression of CD44 was explored in the human protein profiles and is shown in Fig. 4E. Notably, the protein expression level of CD44 in LGG tissue was higher than that in normal brain tissue but lower than that in GBM tissue (Fig. 4E). In summary, these findings strongly suggest that CD44 could be a robust prognostic biomarker for patients with LGGs.
3.5. Astrocytes upregulate U87 cell CD44 expression and facilitate glioma cell viability and migration
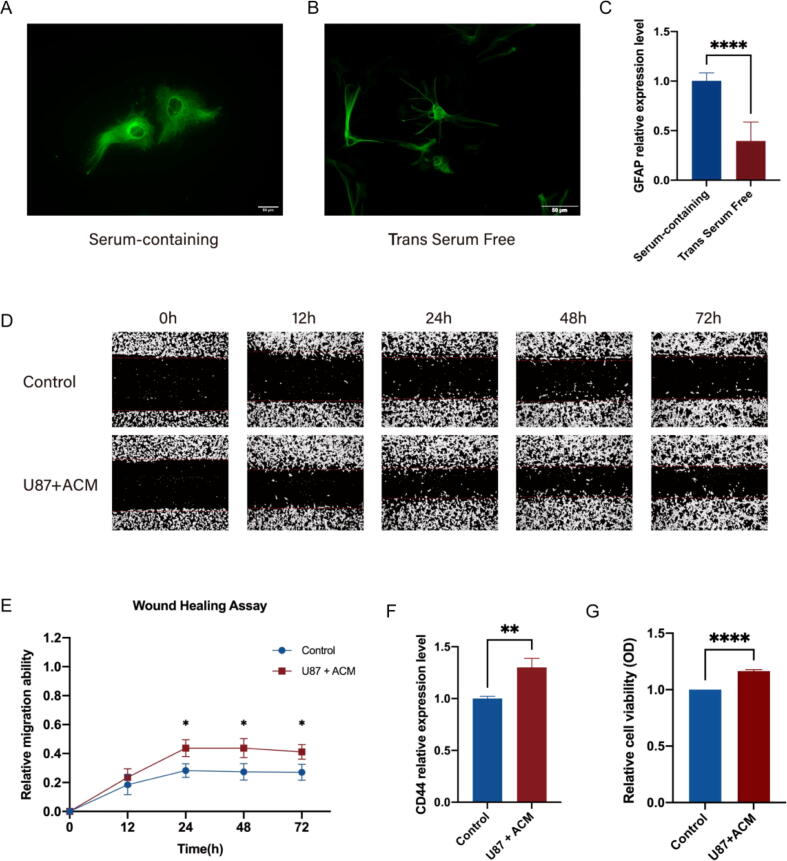
The above analysis indicated that glioma CD44 expression was negatively correlated with LGG patient prognosis. As the first elements to communicate with glioma cells, glial cells are identified to be substantially contributed to tumour growth [48]. Astrocytes haven been described to be actively participated in enhancing GBM cells invasion and proliferation [48], [49]. Given that in the glioma microenvironment, astrocytes may enhance the glioma cell cancer stem cell phenotype via activation of CD44 signalling [18], [19], we exploited trans serum-free astrocytes to reveal the effects of astrocytes on glioma cell CD44 expression and progression under serum-free condition. Compared to astrocytes cultured in serum-containing medium, trans serum-free astrocytes had more processes, better mimicking the morphology of astrocytes in vivo (Fig. 5A, B). After being transferred into serum-free medium for 3 d, the GFAP expression level in astrocytes was significantly decreased, indicating a less reactive state (Fig. 5C). Next, we conducted a wound healing assay and discovered that the migration ability of U87 cells was increased in the U87 + ACM group at 24 h, 48 h and 72 h (Fig. 5D, E). Since the migration ability of U87 cells was already enhanced after culture in ACM for 24 h, we further detected the U87 cell viability and CD44 expression level under ACM stimulation for 24 h. The CD44 expression level and cell viability of U87 cells were both elevated in comparison to those of U87 cells cultured in serum-free medium (Fig. 5F, G). These results indicate that during glioma cell progression, astrocytes may promote glioma cell migration and viability by upregulating CD44 expression.
Fig. 5.
Astrocytes promote migration of the glioma cell line U87 and upregulate CD44 expression. (A) Immunostaining for GFAP. Serum-containing astrocytes were fibroblast-like. (B) Trans serum-free astrocytes had more processes. (C) After transfer into serum-free medium, the GFAP expression level in astrocytes decreased significantly. (D) The effect of ACM on U87 cell migration by a wound healing assay. (E) When cultured in ACM for 24 h, 48 h and 72 h, U87 cells had increased healing ability. (F) RT–qPCR analysis of CD44 expression in U87 cells with or without ACM stimulation. (G) The effect of astrocytes on U87 cell viability after ACM stimulation for 24 h. *: p < 0.05; **: p < 0.01; ****: p < 0.0001; means ± SEMs; n = 3/group. The statistical analysis of RT–qPCR and cell viability assays used an unpaired t test, while migration assays used two-way ANOVA test followed by Šídák's multiple comparisons test.
3.6. CD44 expression was correlated with the proportion of M1 Macrophages in the TME
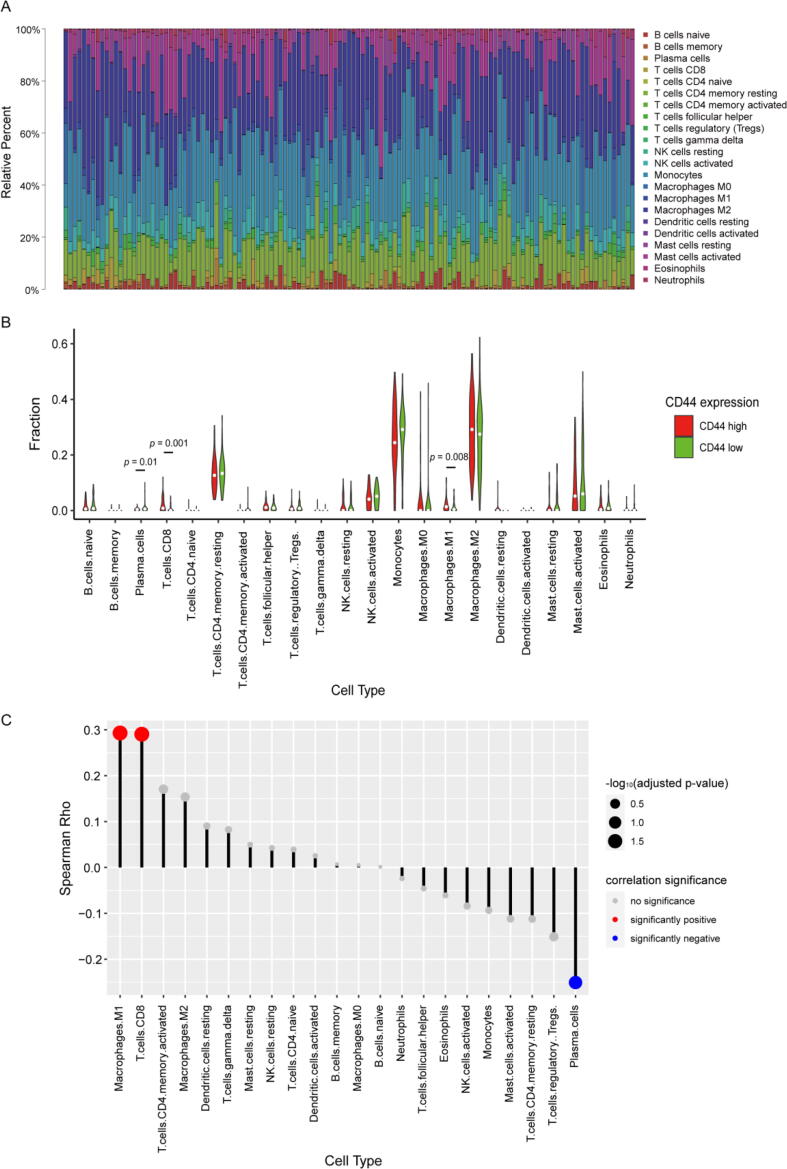
To further decipher the relationship between CD44 expression and the immune microenvironment, CIBERSORT analysis was performed to investigate the ratios of tumour-infiltrating immune subsets in LGG patients in the TCGA database. A total of 22 kinds of immune cell profiles in LGG samples were constructed (Fig. 6A, Fig. S9). The contents of M1 macrophages, CD8+ T cells and plasma cells exhibited significant differences between the high and low CD44 expression groups (Fig. 6B). The results from the lollipop plot showed that M1 macrophages and CD8+ T cells had a positive correlation with CD44 expression, while plasma cells were negatively correlated, suggesting that more M1 macrophages and CD8+ T cells may be recruited into the glioma microenvironment along when the CD44 expression level is elevated in LGGs (Fig. 6C).
Fig. 6.
TIC profile of tumour samples and correlation of TIC proportions with CD44 expression. (A) Bar plot showing the ratios of 22 kinds of TICs in LGG tumour samples. (B) Violin plot displaying the proportions of 22 kinds of immune cells between LGG tumour samples. Low or high CD44 expression was determined compared to the median CD44 expression level. The significance analysis employed the Wilcoxon rank sum test. (C) Lollipop plot showing the correlations of the proportions of 22 kinds of TICs with CD44 expression. The correlation analysis used the Spearman correlation test.
3.7. M1 Macrophages interact with CD44-Positive glioma cells via OPN signalling
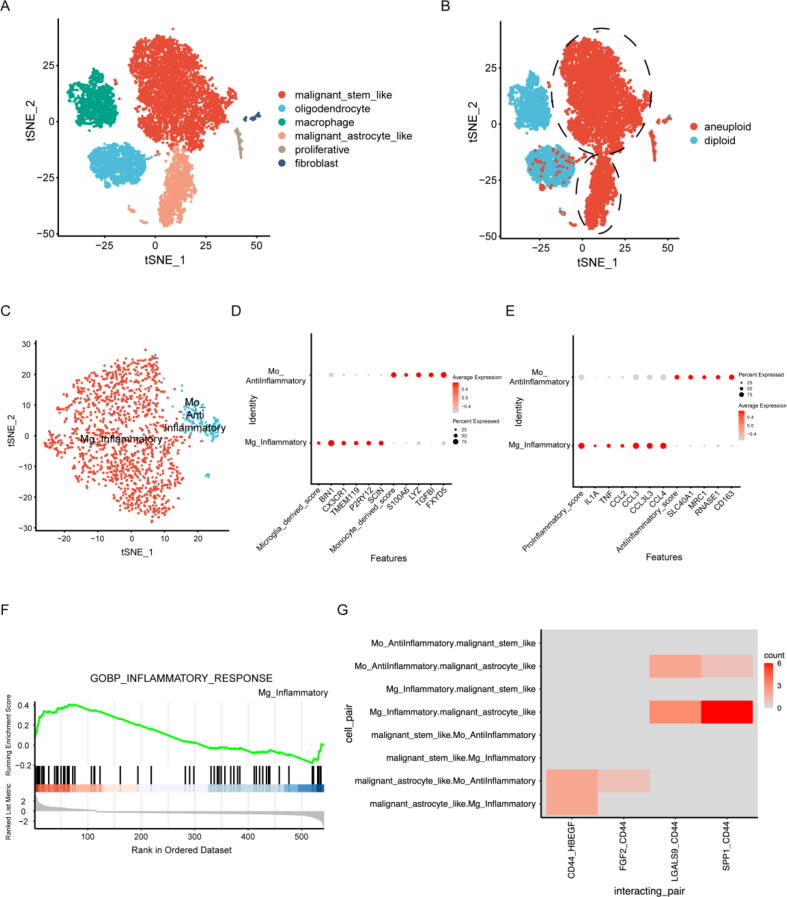
To illustrate the potential interaction and downstream effects of glioma cells and macrophages, previously published single-cell RNA sequencing (scRNA-seq) datasets, including 2 LGG samples from Wang et al. [50] and 8 LGG samples from Benteicher et al. [51], were analysed (Supplemental Table 5). We identified 10,102 cells in total and termed them oligodendrocytes, malignant-astrocyte-like cells, malignant-stem-like cells, macrophages, fibroblasts and proliferating cells (Fig. 7A, S10A) according to the expression of marker genes (Fig. S10B) and the inferred copy number variation status (Fig. 7B). The expression of CD44 was significantly higher in the malignant astrocyte-like cell cluster than in the other clusters (Fig. S10C, Supplemental Table 6). The biological effects of the two clusters of malignant cells were indicated by GSEA (Fig. S11A, B). Malignant-stem-like cells upregulated the expression of genes related to the Hedgehog signalling pathway and Myc targets, while downregulated genes were involved in cholesterol homeostasis, astrocyte differentiation and development, suggesting a low level of differentiation. Astrocyte-like malignant cells upregulated genes related to glial cell differentiation, which suggests their astrocyte-like phenotypes.
Fig. 7.
Single-cell RNA sequencing data analysis reveals cell interactions between macrophages and glioma cells through the OPN-CD44 interaction axis. (A) t-SNE plot of 10,102 cells from 10 LGG astrocytoma samples. Cells are coloured according to their annotated cell identity. (B) Cell aneuploidy predicted by CopyKAT. The circled cell clusters are termed malignant cells. (C) t-SNE plot of 1516 macrophages from 9 samples. (D) Dot plot of marker expression indicating the cell origins of the macrophages. (E) Dot plot of previously reported canonical inflammation-related gene expression for the two macrophage subclusters. (F) The Mg_Inflammatory cell cluster shows enrichment of genes related to the inflammatory response. (G) Significant cell interaction pairs involving CD44 between malignant cells and macrophages in 9 samples as inferred by CellPhoneDB.
To identify subclusters among macrophages, we extracted 1516 macrophages from 9 of 10 samples (as shown in Fig. S9A, MGH103 was excluded because no macrophages were detected in the sample) and reran the clustering process. The cells were clustered into two groups, termed Mg_Inflammatory (M1 phenotype) and Mo_Antiinflammatory (M2 phenotype) (Fig. 7C), based on their inferred cell origins (Fig. 7D) and previously reported canonical inflammation-related gene expression [50], [51], [52], [53] (Fig. 7E). The Mg_Inflammatory cluster of cells highly expressed proinflammatory genes compared to the Mo_AntiInflammatory cluster, as indicated by GSEA (Fig. 7F). Notably, the expression of selenoprotein P (SEPP1) in Mo_anti-inflammatory cells was upregulated (Supplemental Table 7), which was consistent with the finding of a previous study that macrophages with higher SEPP1 expression had an anti-inflammatory phenotype [53]. We analysed possible cell interactions between glioma cells and macrophages. All significant cell interaction pairs between malignant cells and macrophages in 9 samples as inferred by CellPhoneDB was shown in Fig. S11C. The SPP1-CD44 interaction axis between Mg_Inflammatory and malignant-astrocyte-like cells was determined to be significant in 6 out of 9 samples (Fig. 7G, p value < 0.05 and interaction means ≥ 0.5), indicating that such an interaction may be common among LGG patients.
4. Discussion
The TME is essential for cancer growth, metastasis, and response to treatment [54], [55], [56]. The TME in the brain differs remarkably from that in other organs. The blood–brain barrier (BBB) and blood-cerebrospinal fluid barrier, together with the scarcity of avenues to transport antigen-presenting cells into the brain, render the CNS immune privileged [57]. Due to this unique physiological structure, the glioma microenvironment is composed of a large amount of resident and recruited myeloid cells, as well as a comparatively hyporesponsive and exhausted state of tumour-infiltrating lymphocytes [58]. Hence, glioma is known as an immunologically “cold” tumour, for which the microenvironment is immune-suppressive and tumour-permissive [59]. As such, glial cells inhabiting the glioma microenvironment with immunomodulatory functions may play major roles during glioma growth and progression [48]. The TME influences the gene expression of tumour cells and impacts clinical outcomes [60]. Immune and stromal cells are recognized as two major types of nontumor structural components in the TME and have been proposed to be valuable for the diagnostic and prognostic assessment of tumours [55]. Based on these two components, a wide variety of immune-related prognostic markers have been identified and may contribute to prognosis prediction for LGG patients [24], [61].
LGG patients suffer from inevitable recurrence or malignant progression as well as poor prognostic tools and clinical outcomes. Considering the highly heterogeneous nature of LGGs, tumorigenesis, molecular characteristics, therapeutic responses and clinical outcomes vary significantly among LGG patients [62]. Therefore, it is of great importance to seek effective prognostic and therapeutic targets to overcome these challenges. In the current work, we aimed to identify TME-related genes that contribute to the prognosis of LGG patients from the TCGA and CGGA databases. Combining large samples from multiple cohorts with a series of comprehensive bioinformatics analyses, CD44 was identified as a promising prognostic marker for LGG patients. Further experiments indicate that astrocytes may facilitate glioma cell viability and migration by upregulating CD44 expression. CIBERSORT and single-cell RNA sequencing data analysis suggested that M1 macrophages (microglia-derived) may promote the glioma stem-like phenotype by interacting with CD44+ glioma cells via OPN signalling. These findings suggest that during LGG progression, astrocytes enhance CD44 expression in glioma cells, which promotes glioma cell migration and M1 macrophage infiltration and eventually worsens the prognosis of patients with LGGs.
Although the expression of the CD44 gene has been associated with various types of solid tumours, such as non-small cell lung cancer [63], breast cancer [64], and ovarian cancer [65], the prognostic effect of high expression of CD44 for gliomas has been controversial [66], [67]. Some studies claimed that higher tumour expression of CD44 was associated with poor overall survival (OS) in patients with glioma, while others demonstrated no notable association or even the opposite association between these factors [68], [69], [70], [71], [72], [73], [74], [75], [76], [77]. Wu et al. determined that the predictive efficacy of higher CD44 tumour expression for poor OS may be significant in LGG patients but not for patients with glioblastoma [66], [67]. Shortly afterwards, it was confirmed that CD44 was upregulated and was an independent predictor for poor OS in patients with LGGs [78]. CD44-related genes could be prognostic markers and therapeutic targets for LGGs [78]. In the present study, we confirmed that the CD44 gene was one of the DEGs of LGGs and was associated with the clinical outcome of LGG patients. Based on the expression level of CD44, LGG patients could be separated into a low-risk group and a high-risk group, with a significant difference in OS, suggesting the critical prognostic value of CD44 for LGGs.
As a cell membrane glycoprotein, CD44 participates in a number of cellular processes, including cell motility, proliferation, apoptosis and angiogenesis [79]. A previous study showed that compared with that of normal brain tissue, the CD44 expression of human glioma cells is elevated and downregulated CD44 expression ameliorates the migration and invasion of human glioma cells [79]. Meanwhile, as a CSC marker, CD44 expression be used could identify and characterize cancer cells with stemness and aid in improving prognostic evaluation and cancer therapy [12]. Astrocytes have been suggested to actively participate in enhancing GBM cell migration, invasion and proliferation [48], [49]. Notably, astrocytes are proposed to promote glioma stem-like phenotypes via activation of CD44 signalling [18]. In addition, CD44 is not only a panreactive marker for reactive astrocytes but also verified as the distinguishable gene signature between LGGs and GBM in perivascular astrocytes [19], [20]. Coordinated regulation of CD44 expression between astrocytes and glioma cells may occur in different grades of gliomas. Because of the BBB, astrocytes do not contact serum components directly unless they are broken down [41]. Growing evidence indicates that primary astrocytes cultured in serum containing medium exhibit reactive astrocytes phenotype and their morphology are far different from astrocytes in vivo [41], [80]. Moreover, GBM cell line-derived spheres express different levels of CSC markers when cultured in serum-containing medium and serum-free medium [81]. Therefore, to elucidate the effects of astrocytes on glioma CD44 expression under serum-free conditions, we exploited trans serum-free cultured astrocytes. Trans serum-free astrocytes have more processes than astrocytes cultured in serum-containing medium. When astrocytes were transferred into serum-free medium, the reactive marker GFAP was downregulated, indicating the less reactive state of trans serum-free astrocytes. Our experiments confirmed that astrocytes induced glioma U87 cells to express even higher levels of CD44. With the stimulation of astrocytes, U87 cell viability and migration ability increased. Taken together, these data suggested that during glioma cell migration, astrocytes may strengthen their viability and migration ability by upregulating the CD44 expression level of glioma cells during glioma progression.
Aberrant immunomodulation of glial cells has been implicated in gliomas [48]. Studies have shown that GBM-associated astrocytes may induce GBM cells to upregulate periostin and serglycin, which are implicated in the recruitment of M2 GBM-associated macrophages, eventually promoting tumour progression [49], [82], [83]. Next, we wondered whether the astrocyte-induced upregulation of glioma cell CD44 expression would exert effects on the recruitment of macrophages/microglia. High expression of CD44 in human glioma cells, which have been termed mesenchymal-like cells, increases macrophage infiltration and creates an immunosuppressive microenvironment, leading to a poor prognosis [24]. Consistent with this study, our data revealed that the TIC proportion of macrophages varied in LGG patients with different CD44 expression levels. Intriguingly, data on the proportion of TIC in the TME indicated that M1 macrophages and CD8+ T cells were positively correlated with CD44 expression in LGG patients. TAMs are known to produce TGF-β to suppress T cell-mediated tumour clearance [84]. Complex interactions between TAMs and CD8+ T cells contribute to suppressive CD8+ T-cell responses and glioma development [29]. Of note, activated by neuron-derived midkine, CD8+ T cells themselves also induce microglia to secrete Ccl5 to promote LGG cells survival through CD44 binding [85]. Herein, the CD44 receptor of glioma cells may be a crucial downstream factor for the pro-tumorigenic effects of CD8+ T cells on glioma development.
Long thought to be tumoricidal cells, M1 macrophages represent a potential therapeutic strategy for glioma [86], [87]. However, recent studies have highlighted the potential driving effects of M1 macrophages on tumour progression [88], [89]. It has been reported that GSCs not only attract macrophages to tumours and induce M2 polarization but also manage to transdifferentiate M1 macrophages into M2 macrophages, which support GSC maintenance and facilitate tumour growth and progression [89], [90]. After being phagocytosed by M1-TAMs, GSC-derived glioma necrosis products produce IL-12 to support GSCs, which is associated with worse prognosis in recurrent GBMs [91]. Typically, Liu et al. showed that in GBM, pro-inflammatory microglia could secret IL-1β that aids in the proliferation of glioma stem cells [92]. The interaction between the pro-inflammatory macrophages and lower-grade glioma cells via IL-1β was also uncovered in our work in Fig. S11C, revealing that M1 macrophages might be involved across all stages of gliomas progression. To elucidate how M1 macrophages facilitate CD44-positive glioma cell malignancy, single-cell RNA sequencing data were further analysed. It is suggested that microglia-derived brain macrophages, which exhibit the M1 phenotype, may interact with CD44+ glioma cells via OPN signalling. OPN, as a secretory protein, is overexpressed in glioma and essential for GBM stemness and tumorigenicity [93], [94]. An increasing number of studies have identified the crucial roles of OPN in GBM progression [22], [23], [24]. OPN in the normal brain promotes malignant astrocytoma cell migration [95]. It has been reported that glioma cells secrete OPN to recruit M1 and M2 macrophages [23]. Macrophages recruited by glioma cells secrete OPN as well, further facilitating macrophage infiltration and promoting immune suppression and glioma survival [22], [23], [27]. In addition, OPN in the perivascular regions promotes the stem cell-like phenotype of glioma cells via the CD44 intracellular domain, contributing to aggressive glioma growth [18]. Due to the intricate interplay between astrocytes, glioma and M1 macrophages, it stands to reason that as astrocytes elicit the upregulation of glioma CD44 expression, more M1 macrophages are recruited into the tumour niche and promote a glioma stem-like phenotype via OPN-CD44 signalling, ultimately resulting in progressive glioma malignancies.
It is well known that therapeutic resistance is conferred largely by alterations, not in tumour cells but in their environment [10]. A better understanding of the CD44-related prognostic signatures that are involved in shaping the tumour cell extrinsic compartments may be an important step towards the development of more accurate and individualized prognostic factors for LGG patients. The identification of glioma CD44 expression as an orchestrator of glial cell activities in the TME will provide new insights for predicting glioma prognosis, mounting defensive reactions and restoring homeostasis. However, this study has some limitations. First, the biological role of communication between CD44+ glioma and M1 macrophages mediated by the CD44/SPP1 pair should be validated in functional experiments in the near future. Second, we focused only on the prognostic value of CD44 for LGG patients. The prognostic value of CD44 for GBM patients has been reported elsewhere [96]. Whether CD44 is a prognostic marker for all glioma patients has yet to be elucidated. Multicentre studies could be used to further validate the prognostic value of CD44 for glioma patients in future research.
5. Availability of data and materials
The datasets can be downloaded from the UCSC Xena Browser (version 07–19-2019) and CGGA database (https://www.cgga.org.cn). The codes used in the current study are available from the corresponding author.
CRediT authorship contribution statement
Zhanxin Du: Conceptualization, Methodology, Software, Investigation, Formal analysis, Visualization, Validation, Writing – original draft, Writing – review & editing. Yaqing Wang: Conceptualization, Software, Visualization, Writing – review & editing. Jiaqi Liang: Methodology, Software, Data curation, Investigation, Formal analysis, Visualization, Validation. Shaowei Gao: Writing – original draft, Writing – review & editing. Xiaoying Cai: Investigation, Formal analysis. Yu Yu: Investigation, Formal analysis. Zhihui Qi: Investigation, Formal analysis. Jing Li: Conceptualization, Methodology. Yubin Xie: Supervision, Project administration. Zhongxing Wang: Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81601711 and 81971877), the Foundation of Sun Yat-sen University for Young Talent Teachers (Grant No. 19ykzd12) and the Science and Technology Program of Guangzhou, China (Grant No. 201904010418).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.09.003.
Contributor Information
Yubin Xie, Email: xieyb6@mail.sysu.edu.cn.
Zhongxing Wang, Email: wzhxing@mail.sysu.edu.cn, wzx@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lapointe S., Perry A., Butowski N.A. Primary brain tumours in adults. Lancet (London, England) 2018;392(10145):432–446. doi: 10.1016/S0140-6736(18)30990-5. [DOI] [PubMed] [Google Scholar]
- 2.Komori T. Grading of adult diffuse gliomas according to the 2021 WHO Classification of Tumors of the Central Nervous System. Lab Invest. 2021 doi: 10.1038/s41374-021-00667-6. [DOI] [PubMed] [Google Scholar]
- 3.Brat D.J., et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. New Eng J Med. 2015;372(26):2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claus E.B., et al. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus. 2015;38(1):E6. doi: 10.3171/2014.10.FOCUS12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forst D.A., et al. Low-grade gliomas. Oncologist. 2014;19(4):403–413. doi: 10.1634/theoncologist.2013-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Im J.H., et al. Recurrence patterns after maximal surgical resection and postoperative radiotherapy in anaplastic gliomas according to the new 2016 WHO classification. Sci Rep. 2018;8(1):777. doi: 10.1038/s41598-017-19014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wick W., et al. Long-term analysis of the NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with PCV or temozolomide. Neuro Oncol. 2016;18(11):1529–1537. doi: 10.1093/neuonc/now133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumert B.G., et al. Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033–26033): a randomised, open-label, phase 3 intergroup study. LancetOncol. 2016;17(11):1521–1532. doi: 10.1016/S1470-2045(16)30313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagle N., et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29(22):3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meads M.B., Gatenby R.A., Dalton W.S. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer. 2009;9(9):665–674. doi: 10.1038/nrc2714. [DOI] [PubMed] [Google Scholar]
- 11.Junttila M.R., de Sauvage F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501(7467):346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 12.Li W., et al. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci Rep. 2017;7(1):13856. doi: 10.1038/s41598-017-14364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalho J. Cell Reversal From a Differentiated to a Stem-Like State at Cancer Initiation. Front Oncol. 2020;10:541. doi: 10.3389/fonc.2020.00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vieira de Castro J., et al. Exploiting the Complexities of Glioblastoma Stem Cells: Insights for Cancer Initiation and Therapeutic Targeting. Int J Mol Sci. 2020;21(15) doi: 10.3390/ijms21155278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jhaveri N., Chen T.C., Hofman F.M. Tumor vasculature and glioma stem cells: Contributions to glioma progression. Cancer Lett. 2016;380(2):545–551. doi: 10.1016/j.canlet.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 16.Ludwig K., Kornblum H.I. Molecular markers in glioma. J Neurooncol. 2017;134(3):505–512. doi: 10.1007/s11060-017-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudbergsson J.M., et al. Conventional Treatment of Glioblastoma Reveals Persistent CD44 Subpopulations. Mol Neurobiol. 2020;57(9):3943–3955. doi: 10.1007/s12035-020-02004-2. [DOI] [PubMed] [Google Scholar]
- 18.Pietras A., et al. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14(3):357–369. doi: 10.1016/j.stem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz A.M., et al. Astrocyte-specific expression patterns associated with the PDGF-induced glioma microenvironment. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0032453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liddelow S.A., et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541(7638):481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng X., et al. TEM; Trends In Endocrinology and Metabolism: 2022. The membrane receptor CD44: novel insights into metabolism. [DOI] [PubMed] [Google Scholar]
- 22.Ellert-Miklaszewska A., et al. Tumour-processed osteopontin and lactadherin drive the protumorigenic reprogramming of microglia and glioma progression. Oncogene. 2016;35(50):6366–6377. doi: 10.1038/onc.2016.55. [DOI] [PubMed] [Google Scholar]
- 23.Wei J., et al. Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J Clin Investig. 2019;129(1):137–149. doi: 10.1172/JCI121266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo X.Y., et al. Immunogenomic Profiling Demonstrate AC003092.1 as an Immune-Related eRNA in Glioblastoma Multiforme. Front Genet. 2021;12 doi: 10.3389/fgene.2021.633812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szulzewsky F., et al. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0116644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z., et al. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Cancer Res. 2017;77(9):2266–2278. doi: 10.1158/0008-5472.CAN-16-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen P., et al. Symbiotic Macrophage-Glioma Cell Interactions Reveal Synthetic Lethality in PTEN-Null Glioma. Cancer Cell. 2019;35(6) doi: 10.1016/j.ccell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantovani A., et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25(12):677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 29.Tu S., et al. Crosstalk Between Tumor-Associated Microglia/Macrophages and CD8-Positive T Cells Plays a Key Role in Glioblastoma. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshihara K., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman Aaron M., Liu Chih Long, Green Michael R., Gentles Andrew J., Feng Weiguo, Yue Xu., Hoang Chuong D., Diehn Maximilian, Alizadeh Ash A. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldman M.J., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robin X., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B., et al. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics (Oxford, England) 2010;26(4):493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen C., et al. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liberzon A., et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Systems. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin C.-H., et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foo L.C., et al. Development of a method for the purification and culture of rodent astrocytes. Neuron. 2011;71(5):799–811. doi: 10.1016/j.neuron.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCarthy K.D., de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85(3):890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hao Y., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13) doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korsunsky I., et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao R., et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat Biotechnol. 2021;39(5):599–608. doi: 10.1038/s41587-020-00795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Efremova M., et al. Cell PhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15(4):1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 47.The Genotype-Tissue Expression (GTEx) project. Nature Genetics, 2013. 45(6): p. 580-585. [DOI] [PMC free article] [PubMed]
- 48.Parmigiani E., et al. Old Stars and New Players in the Brain Tumor Microenvironment. Front Cell Neurosci. 2021;15 doi: 10.3389/fncel.2021.709917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mega A., et al. Astrocytes enhance glioblastoma growth. Glia. 2020;68(2):316–327. doi: 10.1002/glia.23718. [DOI] [PubMed] [Google Scholar]
- 50.Wang L., et al. The Phenotypes of Proliferating Glioblastoma Cells Reside on a Single Axis of Variation. Cancer Discovery. 2019;9(12):1708–1719. doi: 10.1158/2159-8290.CD-19-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venteicher, A.S., et al., Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science (New York, N.Y.), 2017. 355(6332). [DOI] [PMC free article] [PubMed]
- 52.Zeiner P.S., et al. Distribution and prognostic impact of microglia/macrophage subpopulations in gliomas. Brain Pathology (Zurich, Switzerland) 2019;29(4):513–529. doi: 10.1111/bpa.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pombo Antunes A.R., et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat Neurosci. 2021;24(4):595–610. doi: 10.1038/s41593-020-00789-y. [DOI] [PubMed] [Google Scholar]
- 54.Hanahan D., Coussens L.M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 55.Gieryng A., et al. Immune microenvironment of gliomas. Lab Invest. 2017;97(5):498–518. doi: 10.1038/labinvest.2017.19. [DOI] [PubMed] [Google Scholar]
- 56.Yao M., et al. Astrocytic trans-Differentiation Completes a Multicellular Paracrine Feedback Loop Required for Medulloblastoma Tumor Growth. Cell. 2020;180(3) doi: 10.1016/j.cell.2019.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J., et al. Nanoparticle Drug Delivery System for Glioma and Its Efficacy Improvement Strategies: A Comprehensive Review. Int J Nanomed. 2020;15:2563–2582. doi: 10.2147/IJN.S243223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D'Alessio A., et al. Pathological and Molecular Features of Glioblastoma and Its Peritumoral Tissue. Cancers. 2019;11(4) doi: 10.3390/cancers11040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong N., et al. Tumor Associated Macrophages, as the Dominant Immune Cells, Are an Indispensable Target for Immunologically Cold Tumor-Glioma Therapy? Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.706286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jia D., et al. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging (Albany NY) 2018;10(4):592–605. doi: 10.18632/aging.101415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H., et al. An Immune-Related Signature for Predicting the Prognosis of Lower-Grade Gliomas. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.603341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang T., et al. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016;375(2):263–273. doi: 10.1016/j.canlet.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 63.Zhao S., et al. Prognostic value of CD44 variant exon 6 expression in non-small cell lung cancer: a meta-analysis. Asian Pac J Cancer Prev. 2014;15(16):6761–6766. doi: 10.7314/apjcp.2014.15.16.6761. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z., et al. Prognostic significance of CD24 and CD44 in breast cancer: a meta-analysis. Int J Biol Markers. 2017;32(1):e75–e82. doi: 10.5301/jbm.5000224. [DOI] [PubMed] [Google Scholar]
- 65.Lin J., Ding D. The prognostic role of the cancer stem cell marker CD44 in ovarian cancer: a meta-analysis. Cancer CellInt. 2017;17:8. doi: 10.1186/s12935-016-0376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu G., et al. Expression of CD44 and the survival in glioma: a meta-analysis. Biosci Rep. 2020;40(4) doi: 10.1042/BSR20200520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hou C., et al. Comment on “Expression of CD44 and the survival in glioma: a meta-analysis”. Biosci Rep. 2020;40(10) doi: 10.1042/BSR20202812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alameda F., et al. Prognostic value of stem cell markers in glioblastoma. Biomarkers. 2019;24(7):677–683. doi: 10.1080/1354750X.2019.1652345. [DOI] [PubMed] [Google Scholar]
- 69.Bien-Möller S., et al. Association of Glioblastoma Multiforme Stem Cell Characteristics, Differentiation, and Microglia Marker Genes with Patient Survival. Stem Cells Int. 2018;2018:9628289. doi: 10.1155/2018/9628289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dong Q., et al. Elevated CD44 expression predicts poor prognosis in patients with low-grade glioma. Oncol Lett. 2019;18(4):3698–3704. doi: 10.3892/ol.2019.10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guadagno E., et al. Immunohistochemical expression of stem cell markers CD44 and nestin in glioblastomas: Evaluation of their prognostic significance. Pathol Res Pract. 2016;212(9):825–832. doi: 10.1016/j.prp.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 72.Hou C., et al. Overexpression of CD44 is associated with a poor prognosis in grade II/III gliomas. J Neurooncol. 2019;145(2):201–210. doi: 10.1007/s11060-019-03288-8. [DOI] [PubMed] [Google Scholar]
- 73.Nishikawa M., et al. Significance of Glioma Stem-Like Cells in the Tumor Periphery That Express High Levels of CD44 in Tumor Invasion, Early Progression, and Poor Prognosis in Glioblastoma. Stem Cells Int. 2018;2018:5387041. doi: 10.1155/2018/5387041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pinel B., et al. Mesenchymal subtype of glioblastomas with high DNA-PKcs expression is associated with better response to radiotherapy and temozolomide. J Neurooncol. 2017;132(2):287–294. doi: 10.1007/s11060-016-2367-7. [DOI] [PubMed] [Google Scholar]
- 75.Ranuncolo S.M., et al. CD44 expression in human gliomas. J Surg Oncol. 2002;79(1):30–35. doi: 10.1002/jso.10045. discussion 35–6. [DOI] [PubMed] [Google Scholar]
- 76.Sooman L., et al. FGF2 as a potential prognostic biomarker for proneural glioma patients. Acta Oncol. 2015;54(3):385–394. doi: 10.3109/0284186X.2014.951492. [DOI] [PubMed] [Google Scholar]
- 77.Wei K.C., et al. Evaluation of the prognostic value of CD44 in glioblastoma multiforme. Anticancer Res. 2010;30(1):253–259. [PubMed] [Google Scholar]
- 78.Xiao Y., et al. A Novel Four-Gene Signature Associated With Immune Checkpoint for Predicting Prognosis in Lower-Grade Glioma. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.605737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Okada H., et al. Suppression of CD44 expression decreases migration and invasion of human glioma cells. Int J Cancer. 1996;66(2):255–260. doi: 10.1002/(SICI)1097-0215(19960410)66:2<255::AID-IJC20>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 80.Zamanian J.L., et al. Genomic analysis of reactive astrogliosis. J Neurosci : Off J Soc Neurosci. 2012;32(18):6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hong X., Chedid K., Kalkanis S.N. Glioblastoma cell line-derived spheres in serum-containing medium versus serum-free medium: a comparison of cancer stem cell properties. Int J Oncol. 2012;41(5):1693–1700. doi: 10.3892/ijo.2012.1592. [DOI] [PubMed] [Google Scholar]
- 82.Markovic D.S., et al. Microglia stimulate the invasiveness of glioma cells by increasing the activity of metalloprotease-2. J Neuropathol Exp Neurol. 2005;64(9):754–762. doi: 10.1097/01.jnen.0000178445.33972.a9. [DOI] [PubMed] [Google Scholar]
- 83.Le D.M., et al. Exploitation of astrocytes by glioma cells to facilitate invasiveness: a mechanism involving matrix metalloproteinase-2 and the urokinase-type plasminogen activator-plasmin cascade. J Neurosci : Off J Soc Neurosci. 2003;23(10):4034–4043. doi: 10.1523/JNEUROSCI.23-10-04034.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomas D.A., Massagué J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8(5):369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 85.Guo X., et al. Midkine activation of CD8 T cells establishes a neuron-immune-cancer axis responsible for low-grade glioma growth. Nat Commun. 2020;11(1):2177. doi: 10.1038/s41467-020-15770-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saha D., Martuza R.L., Rabkin S.D. Macrophage Polarization Contributes to Glioblastoma Eradication by Combination Immunovirotherapy and Immune Checkpoint Blockade. Cancer Cell. 2017;32(2):253–267.e5. doi: 10.1016/j.ccell.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van den Bossche W.B.L., et al. Oncolytic virotherapy in glioblastoma patients induces a tumor macrophage phenotypic shift leading to an altered glioblastoma microenvironment. Neuro Oncol. 2018;20(11):1494–1504. doi: 10.1093/neuonc/noy082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jia X.H., et al. Activation of mesenchymal stem cells by macrophages promotes tumor progression through immune suppressive effects. Oncotarget. 2016;7(15):20934–20944. doi: 10.18632/oncotarget.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guo L., et al. Induction of breast cancer stem cells by M1 macrophages through Lin-28B-let-7-HMGA2 axis. Cancer Lett. 2019;452:213–225. doi: 10.1016/j.canlet.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 90.Nusblat L.M., Carroll M.J., Roth C.M. Crosstalk between M2 macrophages and glioma stem cells. CellOncol (Dordr) 2017;40(5):471–482. doi: 10.1007/s13402-017-0337-5. [DOI] [PubMed] [Google Scholar]
- 91.Tabu K., et al. Glioma stem cell (GSC)-derived autoschizis-like products confer GSC niche properties involving M1-like tumor-associated macrophages. Stem Cells. 2020;38(8):921–935. doi: 10.1002/stem.3193. [DOI] [PubMed] [Google Scholar]
- 92.Liu H., et al. Pro-inflammatory and proliferative microglia drive progression of glioblastoma. Cell Reports. 2021;36(11) doi: 10.1016/j.celrep.2021.109718. [DOI] [PubMed] [Google Scholar]
- 93.Lamour V., et al. Targeting osteopontin suppresses glioblastoma stem-like cell character and tumorigenicity in vivo. Int J Cancer. 2015;137(5):1047–1057. doi: 10.1002/ijc.29454. [DOI] [PubMed] [Google Scholar]
- 94.Polat B., et al. Differences in stem cell marker and osteopontin expression in primary and recurrent glioblastoma. Cancer Cell International. 2022;22(1):87. doi: 10.1186/s12935-022-02510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ding Q., et al. Promotion of malignant astrocytoma cell migration by osteopontin expressed in the normal brain: differences in integrin signaling during cell adhesion to osteopontin versus vitronectin. Cancer Res. 2002;62(18):5336–5343. doi: 10.1100/tsw.2002.247. [DOI] [PubMed] [Google Scholar]
- 96.Si D., et al. High Expression of CD44 Predicts a Poor Prognosis in Glioblastomas. Cancer Manage Res. 2020;12:769–775. doi: 10.2147/CMAR.S233423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets can be downloaded from the UCSC Xena Browser (version 07–19-2019) and CGGA database (https://www.cgga.org.cn). The codes used in the current study are available from the corresponding author.